
Drosophila kikkawai (Fruit fly)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Endopterygota; Diptera; Brachycera; Muscomorpha; Eremoneura; Cyclorrhapha; Schizophora; Acalyptratae;
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
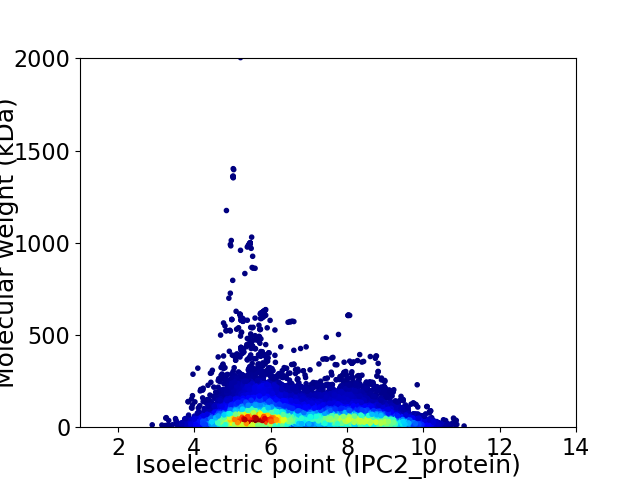
Virtual 2D-PAGE plot for 19992 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6P4I829|A0A6P4I829_DROKI uncharacterized protein LOC108076654 OS=Drosophila kikkawai OX=30033 GN=LOC108076654 PE=4 SV=1
MM1 pKa = 7.2QSKK4 pKa = 10.56LCVVLLALGLSQILAVPAPSLVLTHH29 pKa = 7.18PEE31 pKa = 3.95TNFMQFLMQSRR42 pKa = 11.84DD43 pKa = 3.3LANDD47 pKa = 3.61GTHH50 pKa = 6.71SLEE53 pKa = 4.15CLSYY57 pKa = 8.74YY58 pKa = 10.41TPLLNSAVEE67 pKa = 4.32KK68 pKa = 10.83YY69 pKa = 11.13NNDD72 pKa = 2.84FKK74 pKa = 11.66ACLDD78 pKa = 3.69EE79 pKa = 4.76ATLQVAAINDD89 pKa = 3.66DD90 pKa = 3.64TKK92 pKa = 11.47EE93 pKa = 3.83NRR95 pKa = 11.84TLIDD99 pKa = 3.14QSATDD104 pKa = 3.35SCAALEE110 pKa = 4.41TCSQLTAADD119 pKa = 4.72EE120 pKa = 4.29YY121 pKa = 11.03FEE123 pKa = 5.19CYY125 pKa = 10.21SEE127 pKa = 4.96AGSSNAKK134 pKa = 10.53SMFTISANASEE145 pKa = 4.15LLAYY149 pKa = 10.38VKK151 pKa = 10.2EE152 pKa = 4.15QVHH155 pKa = 6.61IIQVNEE161 pKa = 4.09YY162 pKa = 8.98ICTNKK167 pKa = 8.13TQRR170 pKa = 11.84TYY172 pKa = 11.64AEE174 pKa = 4.21DD175 pKa = 3.3TADD178 pKa = 3.26IYY180 pKa = 11.41EE181 pKa = 4.25SLSLCFSGAPIPTEE195 pKa = 4.11TSSTAAPEE203 pKa = 4.37TSSSLDD209 pKa = 3.59PEE211 pKa = 4.51SSTDD215 pKa = 3.36SSSSEE220 pKa = 4.15PEE222 pKa = 3.93SSSTAEE228 pKa = 4.38PEE230 pKa = 4.2SSSTDD235 pKa = 3.14SSSAEE240 pKa = 4.06PEE242 pKa = 4.39SSSSAAPEE250 pKa = 4.03SSTDD254 pKa = 3.33SSSVEE259 pKa = 4.06PEE261 pKa = 4.13SSSTAAPEE269 pKa = 4.02SSTDD273 pKa = 3.26SSSAEE278 pKa = 4.06PEE280 pKa = 4.39SSSSAAPEE288 pKa = 4.47DD289 pKa = 3.94SSSAEE294 pKa = 4.13PEE296 pKa = 4.39SSSSAAPEE304 pKa = 4.47DD305 pKa = 3.94SSSAEE310 pKa = 4.17PEE312 pKa = 4.32SSSTAAPEE320 pKa = 4.52DD321 pKa = 3.85SSSAEE326 pKa = 4.13PEE328 pKa = 4.39SSSSAAPEE336 pKa = 4.47DD337 pKa = 3.94SSSAEE342 pKa = 4.18PEE344 pKa = 4.17SSSAAPEE351 pKa = 4.08SSTAEE356 pKa = 4.2PEE358 pKa = 4.45SSSSAAPDD366 pKa = 3.52APEE369 pKa = 4.45EE370 pKa = 4.04DD371 pKa = 3.68LKK373 pKa = 11.28QIYY376 pKa = 8.72TGNSNKK382 pKa = 9.77NLQDD386 pKa = 3.18ILKK389 pKa = 10.2NLQTWFKK396 pKa = 7.99TQRR399 pKa = 11.84AA400 pKa = 3.9
MM1 pKa = 7.2QSKK4 pKa = 10.56LCVVLLALGLSQILAVPAPSLVLTHH29 pKa = 7.18PEE31 pKa = 3.95TNFMQFLMQSRR42 pKa = 11.84DD43 pKa = 3.3LANDD47 pKa = 3.61GTHH50 pKa = 6.71SLEE53 pKa = 4.15CLSYY57 pKa = 8.74YY58 pKa = 10.41TPLLNSAVEE67 pKa = 4.32KK68 pKa = 10.83YY69 pKa = 11.13NNDD72 pKa = 2.84FKK74 pKa = 11.66ACLDD78 pKa = 3.69EE79 pKa = 4.76ATLQVAAINDD89 pKa = 3.66DD90 pKa = 3.64TKK92 pKa = 11.47EE93 pKa = 3.83NRR95 pKa = 11.84TLIDD99 pKa = 3.14QSATDD104 pKa = 3.35SCAALEE110 pKa = 4.41TCSQLTAADD119 pKa = 4.72EE120 pKa = 4.29YY121 pKa = 11.03FEE123 pKa = 5.19CYY125 pKa = 10.21SEE127 pKa = 4.96AGSSNAKK134 pKa = 10.53SMFTISANASEE145 pKa = 4.15LLAYY149 pKa = 10.38VKK151 pKa = 10.2EE152 pKa = 4.15QVHH155 pKa = 6.61IIQVNEE161 pKa = 4.09YY162 pKa = 8.98ICTNKK167 pKa = 8.13TQRR170 pKa = 11.84TYY172 pKa = 11.64AEE174 pKa = 4.21DD175 pKa = 3.3TADD178 pKa = 3.26IYY180 pKa = 11.41EE181 pKa = 4.25SLSLCFSGAPIPTEE195 pKa = 4.11TSSTAAPEE203 pKa = 4.37TSSSLDD209 pKa = 3.59PEE211 pKa = 4.51SSTDD215 pKa = 3.36SSSSEE220 pKa = 4.15PEE222 pKa = 3.93SSSTAEE228 pKa = 4.38PEE230 pKa = 4.2SSSTDD235 pKa = 3.14SSSAEE240 pKa = 4.06PEE242 pKa = 4.39SSSSAAPEE250 pKa = 4.03SSTDD254 pKa = 3.33SSSVEE259 pKa = 4.06PEE261 pKa = 4.13SSSTAAPEE269 pKa = 4.02SSTDD273 pKa = 3.26SSSAEE278 pKa = 4.06PEE280 pKa = 4.39SSSSAAPEE288 pKa = 4.47DD289 pKa = 3.94SSSAEE294 pKa = 4.13PEE296 pKa = 4.39SSSSAAPEE304 pKa = 4.47DD305 pKa = 3.94SSSAEE310 pKa = 4.17PEE312 pKa = 4.32SSSTAAPEE320 pKa = 4.52DD321 pKa = 3.85SSSAEE326 pKa = 4.13PEE328 pKa = 4.39SSSSAAPEE336 pKa = 4.47DD337 pKa = 3.94SSSAEE342 pKa = 4.18PEE344 pKa = 4.17SSSAAPEE351 pKa = 4.08SSTAEE356 pKa = 4.2PEE358 pKa = 4.45SSSSAAPDD366 pKa = 3.52APEE369 pKa = 4.45EE370 pKa = 4.04DD371 pKa = 3.68LKK373 pKa = 11.28QIYY376 pKa = 8.72TGNSNKK382 pKa = 9.77NLQDD386 pKa = 3.18ILKK389 pKa = 10.2NLQTWFKK396 pKa = 7.99TQRR399 pKa = 11.84AA400 pKa = 3.9
Molecular weight: 41.75 kDa
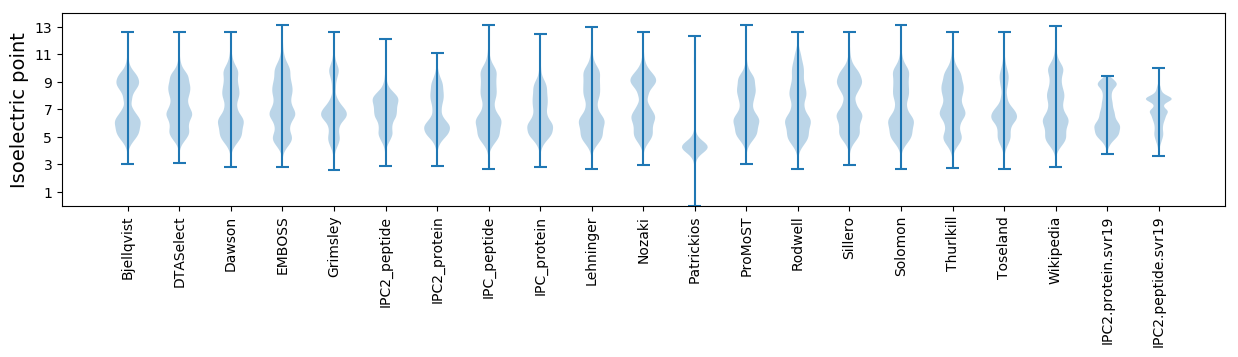
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6P4IPG3|A0A6P4IPG3_DROKI Cell division protein kinase 7 OS=Drosophila kikkawai OX=30033 GN=LOC108080235 PE=3 SV=1
MM1 pKa = 7.86RR2 pKa = 11.84LLLAIVGCLFCALFGQTSGTPEE24 pKa = 4.13CRR26 pKa = 11.84PPLVILPKK34 pKa = 10.01PDD36 pKa = 3.07INSRR40 pKa = 11.84RR41 pKa = 11.84ARR43 pKa = 11.84PVIRR47 pKa = 11.84PLPRR51 pKa = 11.84PPPPKK56 pKa = 9.99VVRR59 pKa = 11.84PPVKK63 pKa = 10.12PPIKK67 pKa = 9.88RR68 pKa = 11.84PPIVPPRR75 pKa = 11.84ITRR78 pKa = 11.84RR79 pKa = 11.84PITPPRR85 pKa = 11.84ITRR88 pKa = 11.84RR89 pKa = 11.84PPIPPPRR96 pKa = 11.84ITRR99 pKa = 11.84RR100 pKa = 11.84PPIQPPRR107 pKa = 11.84ITRR110 pKa = 11.84RR111 pKa = 11.84PPIPPVTRR119 pKa = 11.84KK120 pKa = 7.21PTRR123 pKa = 11.84PPVTQRR129 pKa = 11.84ATAKK133 pKa = 8.87ATTRR137 pKa = 11.84KK138 pKa = 9.36FLIWSKK144 pKa = 10.78SPTRR148 pKa = 11.84RR149 pKa = 11.84VPTRR153 pKa = 11.84PNIRR157 pKa = 11.84PTLWTRR163 pKa = 11.84PNIRR167 pKa = 11.84PTRR170 pKa = 11.84PPVRR174 pKa = 11.84PPSPTVRR181 pKa = 11.84PTVPRR186 pKa = 11.84VRR188 pKa = 11.84PTLWTRR194 pKa = 11.84PPVRR198 pKa = 11.84VTVRR202 pKa = 11.84PTNRR206 pKa = 11.84APVTRR211 pKa = 11.84PPVRR215 pKa = 11.84PTNRR219 pKa = 11.84ATLWTRR225 pKa = 11.84NPMGTTRR232 pKa = 11.84PPRR235 pKa = 11.84PTMKK239 pKa = 10.26PSRR242 pKa = 11.84PTLWTRR248 pKa = 11.84SPTRR252 pKa = 11.84STQQTTRR259 pKa = 11.84HH260 pKa = 4.92LTFIFEE266 pKa = 4.45GSSAAPVTPMDD277 pKa = 3.67VTEE280 pKa = 5.1DD281 pKa = 3.68PQITRR286 pKa = 11.84TTKK289 pKa = 9.96SLIFIFPRR297 pKa = 11.84DD298 pKa = 3.65TTAMARR304 pKa = 11.84GPNWRR309 pKa = 11.84VHH311 pKa = 4.35PTGRR315 pKa = 11.84PRR317 pKa = 11.84RR318 pKa = 11.84VTMVSS323 pKa = 2.91
MM1 pKa = 7.86RR2 pKa = 11.84LLLAIVGCLFCALFGQTSGTPEE24 pKa = 4.13CRR26 pKa = 11.84PPLVILPKK34 pKa = 10.01PDD36 pKa = 3.07INSRR40 pKa = 11.84RR41 pKa = 11.84ARR43 pKa = 11.84PVIRR47 pKa = 11.84PLPRR51 pKa = 11.84PPPPKK56 pKa = 9.99VVRR59 pKa = 11.84PPVKK63 pKa = 10.12PPIKK67 pKa = 9.88RR68 pKa = 11.84PPIVPPRR75 pKa = 11.84ITRR78 pKa = 11.84RR79 pKa = 11.84PITPPRR85 pKa = 11.84ITRR88 pKa = 11.84RR89 pKa = 11.84PPIPPPRR96 pKa = 11.84ITRR99 pKa = 11.84RR100 pKa = 11.84PPIQPPRR107 pKa = 11.84ITRR110 pKa = 11.84RR111 pKa = 11.84PPIPPVTRR119 pKa = 11.84KK120 pKa = 7.21PTRR123 pKa = 11.84PPVTQRR129 pKa = 11.84ATAKK133 pKa = 8.87ATTRR137 pKa = 11.84KK138 pKa = 9.36FLIWSKK144 pKa = 10.78SPTRR148 pKa = 11.84RR149 pKa = 11.84VPTRR153 pKa = 11.84PNIRR157 pKa = 11.84PTLWTRR163 pKa = 11.84PNIRR167 pKa = 11.84PTRR170 pKa = 11.84PPVRR174 pKa = 11.84PPSPTVRR181 pKa = 11.84PTVPRR186 pKa = 11.84VRR188 pKa = 11.84PTLWTRR194 pKa = 11.84PPVRR198 pKa = 11.84VTVRR202 pKa = 11.84PTNRR206 pKa = 11.84APVTRR211 pKa = 11.84PPVRR215 pKa = 11.84PTNRR219 pKa = 11.84ATLWTRR225 pKa = 11.84NPMGTTRR232 pKa = 11.84PPRR235 pKa = 11.84PTMKK239 pKa = 10.26PSRR242 pKa = 11.84PTLWTRR248 pKa = 11.84SPTRR252 pKa = 11.84STQQTTRR259 pKa = 11.84HH260 pKa = 4.92LTFIFEE266 pKa = 4.45GSSAAPVTPMDD277 pKa = 3.67VTEE280 pKa = 5.1DD281 pKa = 3.68PQITRR286 pKa = 11.84TTKK289 pKa = 9.96SLIFIFPRR297 pKa = 11.84DD298 pKa = 3.65TTAMARR304 pKa = 11.84GPNWRR309 pKa = 11.84VHH311 pKa = 4.35PTGRR315 pKa = 11.84PRR317 pKa = 11.84RR318 pKa = 11.84VTMVSS323 pKa = 2.91
Molecular weight: 36.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
12791145 |
32 |
18810 |
639.8 |
71.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.849 ± 0.02 | 1.72 ± 0.024 |
5.156 ± 0.014 | 6.542 ± 0.025 |
3.308 ± 0.014 | 6.354 ± 0.023 |
2.655 ± 0.01 | 4.665 ± 0.014 |
5.415 ± 0.018 | 9.048 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.283 ± 0.009 | 4.589 ± 0.012 |
5.626 ± 0.024 | 5.54 ± 0.027 |
5.562 ± 0.015 | 8.44 ± 0.027 |
5.688 ± 0.014 | 5.796 ± 0.015 |
0.956 ± 0.005 | 2.806 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
