
Xenococcus sp. PCC 7305
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Pleurocapsales; Xenococcaceae; Xenococcus; unclassified Xenococcus
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
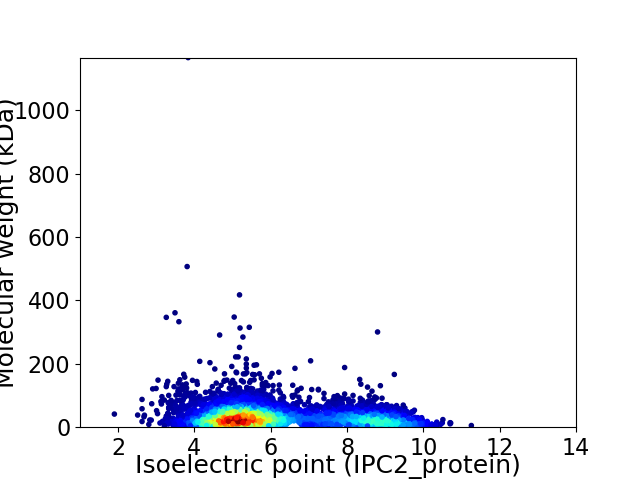
Virtual 2D-PAGE plot for 5347 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L8M053|L8M053_9CYAN Uncharacterized protein OS=Xenococcus sp. PCC 7305 OX=102125 GN=Xen7305DRAFT_00004360 PE=4 SV=1
MM1 pKa = 7.78AEE3 pKa = 4.33NLIMWIKK10 pKa = 10.49QLGSPEE16 pKa = 4.21NDD18 pKa = 2.98LSQGIAIDD26 pKa = 3.66SNGNVFISGSTRR38 pKa = 11.84GTLGNTNFGNSDD50 pKa = 3.24TWVAKK55 pKa = 10.27YY56 pKa = 10.29DD57 pKa = 3.68HH58 pKa = 6.6QGNLLWTQQFGTSNLDD74 pKa = 3.45DD75 pKa = 4.4FEE77 pKa = 6.4GLATDD82 pKa = 3.86INGHH86 pKa = 5.21VYY88 pKa = 10.16IAGSSLGTIEE98 pKa = 4.25GTFQIDD104 pKa = 2.6SDD106 pKa = 3.77AWVNKK111 pKa = 10.15YY112 pKa = 10.33DD113 pKa = 4.06GQGNLLWSQQLGFSGFDD130 pKa = 3.13SATGVATDD138 pKa = 4.22LGGNVYY144 pKa = 10.34ISGFTNGISDD154 pKa = 3.75EE155 pKa = 4.4ANGGGSVAWVTKK167 pKa = 10.75YY168 pKa = 10.62DD169 pKa = 3.66VQGNLLWNQQLDD181 pKa = 3.63ASGSEE186 pKa = 3.61ISAGVTTDD194 pKa = 2.43IHH196 pKa = 6.79GNFYY200 pKa = 10.56ISGDD204 pKa = 3.68TTRR207 pKa = 11.84SNSEE211 pKa = 3.66NTNAWVAKK219 pKa = 9.92YY220 pKa = 9.99DD221 pKa = 4.29AEE223 pKa = 5.91GNLLWNQQLDD233 pKa = 3.8SAEE236 pKa = 4.34AEE238 pKa = 3.95FDD240 pKa = 3.44YY241 pKa = 11.2SQAVTTDD248 pKa = 3.29LSGNVYY254 pKa = 10.18IAGGTNGSIDD264 pKa = 3.64RR265 pKa = 11.84GNAGLNDD272 pKa = 3.44AWVAKK277 pKa = 10.53YY278 pKa = 10.48DD279 pKa = 4.28GEE281 pKa = 5.22GNLLWTEE288 pKa = 3.9QLGTVGNDD296 pKa = 2.85AAEE299 pKa = 5.22GITTDD304 pKa = 3.7SEE306 pKa = 4.78GNVYY310 pKa = 10.33ISGFTNEE317 pKa = 4.45TLGEE321 pKa = 4.13ANAGGSDD328 pKa = 3.21AWVAKK333 pKa = 10.31YY334 pKa = 10.59DD335 pKa = 4.3SNGNLLWTEE344 pKa = 4.01QFGSPEE350 pKa = 4.52DD351 pKa = 4.62DD352 pKa = 4.11DD353 pKa = 4.64SQGVAIDD360 pKa = 4.28SNGSLYY366 pKa = 10.9LSGSTEE372 pKa = 4.07GNLDD376 pKa = 3.36GVNFGGEE383 pKa = 4.4DD384 pKa = 2.94AWVAQISQPIIPDD397 pKa = 3.17NGGNGGNNGNGGGSAPLLTTPINRR421 pKa = 11.84FQNNSLPGTYY431 pKa = 10.26LFAGEE436 pKa = 5.27DD437 pKa = 3.62EE438 pKa = 4.63SQGIRR443 pKa = 11.84ANFPNFVEE451 pKa = 4.28EE452 pKa = 4.42GQAFRR457 pKa = 11.84VGVEE461 pKa = 4.07PGDD464 pKa = 3.37NLIRR468 pKa = 11.84LNRR471 pKa = 11.84FQNSNVPGTYY481 pKa = 9.86LYY483 pKa = 10.85AGEE486 pKa = 4.96EE487 pKa = 4.24EE488 pKa = 4.52SQSIRR493 pKa = 11.84ANFSNFIEE501 pKa = 4.29EE502 pKa = 4.7GIAFYY507 pKa = 10.94VYY509 pKa = 10.78AGTADD514 pKa = 3.12IGTDD518 pKa = 3.66FYY520 pKa = 11.4RR521 pKa = 11.84FQNQNVPGTYY531 pKa = 9.43IFVGGNEE538 pKa = 4.11RR539 pKa = 11.84QNILANFPHH548 pKa = 6.09FHH550 pKa = 6.78EE551 pKa = 4.79EE552 pKa = 4.45GIAFEE557 pKa = 4.46VGTT560 pKa = 4.42
MM1 pKa = 7.78AEE3 pKa = 4.33NLIMWIKK10 pKa = 10.49QLGSPEE16 pKa = 4.21NDD18 pKa = 2.98LSQGIAIDD26 pKa = 3.66SNGNVFISGSTRR38 pKa = 11.84GTLGNTNFGNSDD50 pKa = 3.24TWVAKK55 pKa = 10.27YY56 pKa = 10.29DD57 pKa = 3.68HH58 pKa = 6.6QGNLLWTQQFGTSNLDD74 pKa = 3.45DD75 pKa = 4.4FEE77 pKa = 6.4GLATDD82 pKa = 3.86INGHH86 pKa = 5.21VYY88 pKa = 10.16IAGSSLGTIEE98 pKa = 4.25GTFQIDD104 pKa = 2.6SDD106 pKa = 3.77AWVNKK111 pKa = 10.15YY112 pKa = 10.33DD113 pKa = 4.06GQGNLLWSQQLGFSGFDD130 pKa = 3.13SATGVATDD138 pKa = 4.22LGGNVYY144 pKa = 10.34ISGFTNGISDD154 pKa = 3.75EE155 pKa = 4.4ANGGGSVAWVTKK167 pKa = 10.75YY168 pKa = 10.62DD169 pKa = 3.66VQGNLLWNQQLDD181 pKa = 3.63ASGSEE186 pKa = 3.61ISAGVTTDD194 pKa = 2.43IHH196 pKa = 6.79GNFYY200 pKa = 10.56ISGDD204 pKa = 3.68TTRR207 pKa = 11.84SNSEE211 pKa = 3.66NTNAWVAKK219 pKa = 9.92YY220 pKa = 9.99DD221 pKa = 4.29AEE223 pKa = 5.91GNLLWNQQLDD233 pKa = 3.8SAEE236 pKa = 4.34AEE238 pKa = 3.95FDD240 pKa = 3.44YY241 pKa = 11.2SQAVTTDD248 pKa = 3.29LSGNVYY254 pKa = 10.18IAGGTNGSIDD264 pKa = 3.64RR265 pKa = 11.84GNAGLNDD272 pKa = 3.44AWVAKK277 pKa = 10.53YY278 pKa = 10.48DD279 pKa = 4.28GEE281 pKa = 5.22GNLLWTEE288 pKa = 3.9QLGTVGNDD296 pKa = 2.85AAEE299 pKa = 5.22GITTDD304 pKa = 3.7SEE306 pKa = 4.78GNVYY310 pKa = 10.33ISGFTNEE317 pKa = 4.45TLGEE321 pKa = 4.13ANAGGSDD328 pKa = 3.21AWVAKK333 pKa = 10.31YY334 pKa = 10.59DD335 pKa = 4.3SNGNLLWTEE344 pKa = 4.01QFGSPEE350 pKa = 4.52DD351 pKa = 4.62DD352 pKa = 4.11DD353 pKa = 4.64SQGVAIDD360 pKa = 4.28SNGSLYY366 pKa = 10.9LSGSTEE372 pKa = 4.07GNLDD376 pKa = 3.36GVNFGGEE383 pKa = 4.4DD384 pKa = 2.94AWVAQISQPIIPDD397 pKa = 3.17NGGNGGNNGNGGGSAPLLTTPINRR421 pKa = 11.84FQNNSLPGTYY431 pKa = 10.26LFAGEE436 pKa = 5.27DD437 pKa = 3.62EE438 pKa = 4.63SQGIRR443 pKa = 11.84ANFPNFVEE451 pKa = 4.28EE452 pKa = 4.42GQAFRR457 pKa = 11.84VGVEE461 pKa = 4.07PGDD464 pKa = 3.37NLIRR468 pKa = 11.84LNRR471 pKa = 11.84FQNSNVPGTYY481 pKa = 9.86LYY483 pKa = 10.85AGEE486 pKa = 4.96EE487 pKa = 4.24EE488 pKa = 4.52SQSIRR493 pKa = 11.84ANFSNFIEE501 pKa = 4.29EE502 pKa = 4.7GIAFYY507 pKa = 10.94VYY509 pKa = 10.78AGTADD514 pKa = 3.12IGTDD518 pKa = 3.66FYY520 pKa = 11.4RR521 pKa = 11.84FQNQNVPGTYY531 pKa = 9.43IFVGGNEE538 pKa = 4.11RR539 pKa = 11.84QNILANFPHH548 pKa = 6.09FHH550 pKa = 6.78EE551 pKa = 4.79EE552 pKa = 4.45GIAFEE557 pKa = 4.46VGTT560 pKa = 4.42
Molecular weight: 59.7 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L8LYW6|L8LYW6_9CYAN Uncharacterized protein OS=Xenococcus sp. PCC 7305 OX=102125 GN=Xen7305DRAFT_00009040 PE=4 SV=1
MM1 pKa = 7.55TKK3 pKa = 9.01RR4 pKa = 11.84TLGGTSRR11 pKa = 11.84KK12 pKa = 7.83QKK14 pKa = 9.77RR15 pKa = 11.84KK16 pKa = 8.61SGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TKK26 pKa = 10.37NGRR29 pKa = 11.84KK30 pKa = 9.19VIQARR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.98KK38 pKa = 9.65GRR40 pKa = 11.84HH41 pKa = 5.0RR42 pKa = 11.84LSVV45 pKa = 3.12
MM1 pKa = 7.55TKK3 pKa = 9.01RR4 pKa = 11.84TLGGTSRR11 pKa = 11.84KK12 pKa = 7.83QKK14 pKa = 9.77RR15 pKa = 11.84KK16 pKa = 8.61SGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TKK26 pKa = 10.37NGRR29 pKa = 11.84KK30 pKa = 9.19VIQARR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.98KK38 pKa = 9.65GRR40 pKa = 11.84HH41 pKa = 5.0RR42 pKa = 11.84LSVV45 pKa = 3.12
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1695650 |
20 |
10636 |
317.1 |
35.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.621 ± 0.036 | 0.928 ± 0.014 |
5.477 ± 0.053 | 6.55 ± 0.033 |
4.161 ± 0.025 | 6.652 ± 0.054 |
1.682 ± 0.018 | 7.544 ± 0.029 |
5.289 ± 0.044 | 10.763 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.719 ± 0.021 | 5.029 ± 0.036 |
4.196 ± 0.024 | 5.055 ± 0.035 |
4.518 ± 0.031 | 6.711 ± 0.026 |
5.527 ± 0.038 | 6.074 ± 0.029 |
1.34 ± 0.015 | 3.162 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
