
Enterorhabdus caecimuris B7
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Coriobacteriia; Eggerthellales; Eggerthellaceae; Adlercreutzia; Adlercreutzia caecimuris
Average proteome isoelectric point is 5.78
Get precalculated fractions of proteins
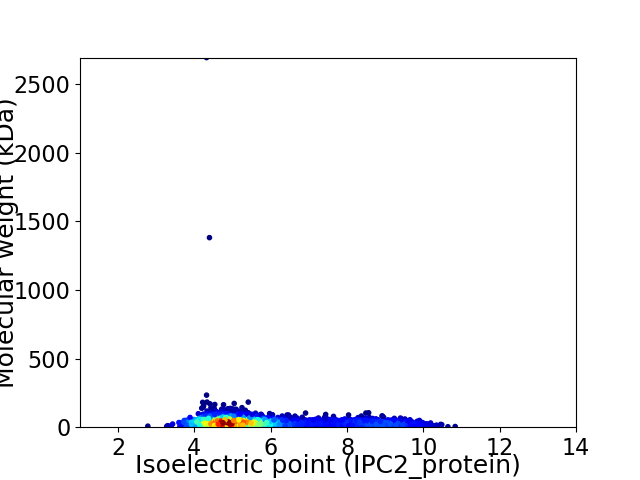
Virtual 2D-PAGE plot for 2435 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
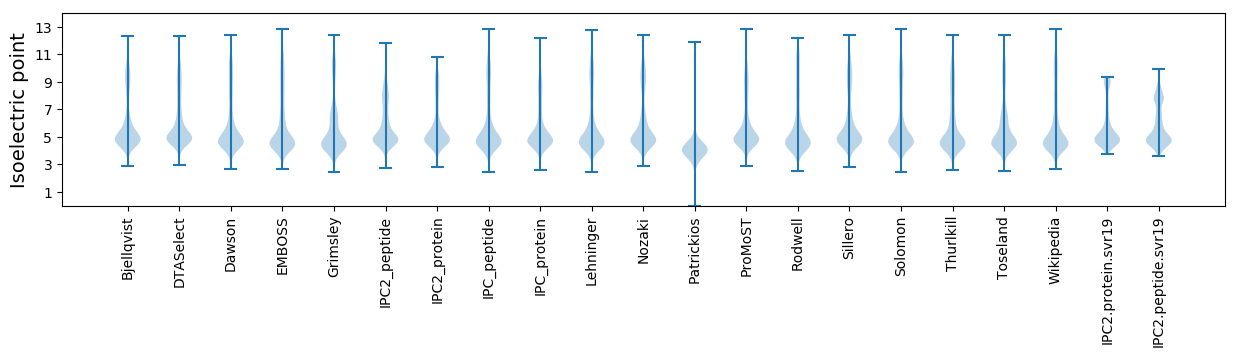
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R9KVX0|R9KVX0_9ACTN YggT family protein OS=Enterorhabdus caecimuris B7 OX=1235794 GN=C811_00935 PE=3 SV=1
MM1 pKa = 7.58RR2 pKa = 11.84MKK4 pKa = 10.61PILAMLLGAALLPISACSGYY24 pKa = 10.4DD25 pKa = 3.07IPEE28 pKa = 4.3EE29 pKa = 4.26VPAPAQSDD37 pKa = 3.27ASNPFEE43 pKa = 4.37EE44 pKa = 4.79AQDD47 pKa = 4.13DD48 pKa = 4.16ASLMAMLSHH57 pKa = 6.84EE58 pKa = 4.45AANPPLSDD66 pKa = 3.23SGEE69 pKa = 4.09RR70 pKa = 11.84EE71 pKa = 3.87PFPYY75 pKa = 10.37DD76 pKa = 3.05GGEE79 pKa = 3.62FRR81 pKa = 11.84LDD83 pKa = 3.23YY84 pKa = 10.38RR85 pKa = 11.84FSVSGDD91 pKa = 3.24LDD93 pKa = 3.69SVGFLLFLDD102 pKa = 5.48GKK104 pKa = 7.77PQPYY108 pKa = 10.08RR109 pKa = 11.84IDD111 pKa = 3.89GQSSDD116 pKa = 3.73YY117 pKa = 11.21AFCHH121 pKa = 5.93TFDD124 pKa = 3.81AAEE127 pKa = 4.4DD128 pKa = 3.51QDD130 pKa = 3.62IAFLFEE136 pKa = 4.2PVAGHH141 pKa = 7.09SGDD144 pKa = 3.79TLNLTIVSITNPDD157 pKa = 3.6FQPDD161 pKa = 3.68MEE163 pKa = 4.65STSSYY168 pKa = 10.01GWYY171 pKa = 10.3HH172 pKa = 6.12NALSASVEE180 pKa = 4.0MRR182 pKa = 11.84FNADD186 pKa = 3.66PPSSAATAPNPVEE199 pKa = 4.73AFSSCSVHH207 pKa = 5.54EE208 pKa = 4.43EE209 pKa = 4.25KK210 pKa = 10.0VTSRR214 pKa = 11.84YY215 pKa = 10.05LEE217 pKa = 4.05NDD219 pKa = 3.53LAQAGWPDD227 pKa = 3.06ITMDD231 pKa = 3.77TLDD234 pKa = 3.98DD235 pKa = 4.74GIYY238 pKa = 9.07YY239 pKa = 9.56TLTLDD244 pKa = 4.0GDD246 pKa = 4.44TVFDD250 pKa = 4.29NLPLTEE256 pKa = 4.07PVAVRR261 pKa = 11.84FTLCGTPGARR271 pKa = 11.84YY272 pKa = 9.78NISFFLDD279 pKa = 3.67HH280 pKa = 6.82QPMAVNGEE288 pKa = 3.91ASSAVEE294 pKa = 3.74LSKK297 pKa = 11.47GGVAVVEE304 pKa = 4.4GVIDD308 pKa = 4.01PSLLGKK314 pKa = 10.62LSTFYY319 pKa = 10.87CVAVPADD326 pKa = 3.52NAGAPFFKK334 pKa = 10.37TNSILLFQEE343 pKa = 4.22AA344 pKa = 4.03
MM1 pKa = 7.58RR2 pKa = 11.84MKK4 pKa = 10.61PILAMLLGAALLPISACSGYY24 pKa = 10.4DD25 pKa = 3.07IPEE28 pKa = 4.3EE29 pKa = 4.26VPAPAQSDD37 pKa = 3.27ASNPFEE43 pKa = 4.37EE44 pKa = 4.79AQDD47 pKa = 4.13DD48 pKa = 4.16ASLMAMLSHH57 pKa = 6.84EE58 pKa = 4.45AANPPLSDD66 pKa = 3.23SGEE69 pKa = 4.09RR70 pKa = 11.84EE71 pKa = 3.87PFPYY75 pKa = 10.37DD76 pKa = 3.05GGEE79 pKa = 3.62FRR81 pKa = 11.84LDD83 pKa = 3.23YY84 pKa = 10.38RR85 pKa = 11.84FSVSGDD91 pKa = 3.24LDD93 pKa = 3.69SVGFLLFLDD102 pKa = 5.48GKK104 pKa = 7.77PQPYY108 pKa = 10.08RR109 pKa = 11.84IDD111 pKa = 3.89GQSSDD116 pKa = 3.73YY117 pKa = 11.21AFCHH121 pKa = 5.93TFDD124 pKa = 3.81AAEE127 pKa = 4.4DD128 pKa = 3.51QDD130 pKa = 3.62IAFLFEE136 pKa = 4.2PVAGHH141 pKa = 7.09SGDD144 pKa = 3.79TLNLTIVSITNPDD157 pKa = 3.6FQPDD161 pKa = 3.68MEE163 pKa = 4.65STSSYY168 pKa = 10.01GWYY171 pKa = 10.3HH172 pKa = 6.12NALSASVEE180 pKa = 4.0MRR182 pKa = 11.84FNADD186 pKa = 3.66PPSSAATAPNPVEE199 pKa = 4.73AFSSCSVHH207 pKa = 5.54EE208 pKa = 4.43EE209 pKa = 4.25KK210 pKa = 10.0VTSRR214 pKa = 11.84YY215 pKa = 10.05LEE217 pKa = 4.05NDD219 pKa = 3.53LAQAGWPDD227 pKa = 3.06ITMDD231 pKa = 3.77TLDD234 pKa = 3.98DD235 pKa = 4.74GIYY238 pKa = 9.07YY239 pKa = 9.56TLTLDD244 pKa = 4.0GDD246 pKa = 4.44TVFDD250 pKa = 4.29NLPLTEE256 pKa = 4.07PVAVRR261 pKa = 11.84FTLCGTPGARR271 pKa = 11.84YY272 pKa = 9.78NISFFLDD279 pKa = 3.67HH280 pKa = 6.82QPMAVNGEE288 pKa = 3.91ASSAVEE294 pKa = 3.74LSKK297 pKa = 11.47GGVAVVEE304 pKa = 4.4GVIDD308 pKa = 4.01PSLLGKK314 pKa = 10.62LSTFYY319 pKa = 10.87CVAVPADD326 pKa = 3.52NAGAPFFKK334 pKa = 10.37TNSILLFQEE343 pKa = 4.22AA344 pKa = 4.03
Molecular weight: 37.05 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R9LDB6|R9LDB6_9ACTN Dimethylsulfoxide reductase chain B OS=Enterorhabdus caecimuris B7 OX=1235794 GN=C811_00036 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 10.25AAGGRR7 pKa = 11.84GGRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84TPYY15 pKa = 8.01NTPMRR20 pKa = 11.84IFLYY24 pKa = 10.41GISALSWWLSAPATPGEE41 pKa = 4.26GNRR44 pKa = 11.84VGQNALRR51 pKa = 11.84NCKK54 pKa = 9.22PDD56 pKa = 3.34AAAIQYY62 pKa = 8.16LAHH65 pKa = 7.2CCPQIPRR72 pKa = 11.84PYY74 pKa = 9.35HH75 pKa = 4.31VTVSVRR81 pKa = 11.84MKK83 pKa = 10.12RR84 pKa = 11.84CPDD87 pKa = 2.72GVAPHH92 pKa = 7.15RR93 pKa = 11.84SQFKK97 pKa = 9.78LVGRR101 pKa = 11.84PFCRR105 pKa = 11.84VASGIYY111 pKa = 9.83APCPEE116 pKa = 4.34LCFVQVANNLDD127 pKa = 3.37LHH129 pKa = 6.26EE130 pKa = 4.35LVKK133 pKa = 10.96VGNALCGTFFIDD145 pKa = 3.68PKK147 pKa = 10.86ARR149 pKa = 11.84NGLGKK154 pKa = 10.04RR155 pKa = 11.84RR156 pKa = 11.84PLTSVRR162 pKa = 11.84RR163 pKa = 11.84IGAFIEE169 pKa = 4.3RR170 pKa = 11.84NPGILGAKK178 pKa = 7.85PARR181 pKa = 11.84RR182 pKa = 11.84ALGLMVDD189 pKa = 4.94GAASPPEE196 pKa = 4.25VFLAMALGLPYY207 pKa = 10.56RR208 pKa = 11.84FGGYY212 pKa = 7.66QLPGIAANRR221 pKa = 11.84RR222 pKa = 11.84IKK224 pKa = 10.46PSSKK228 pKa = 10.12ARR230 pKa = 11.84AIAHH234 pKa = 7.08RR235 pKa = 11.84STLVPDD241 pKa = 3.95ILCEE245 pKa = 4.04SSRR248 pKa = 11.84LDD250 pKa = 3.75IEE252 pKa = 4.32YY253 pKa = 10.72DD254 pKa = 3.61SNTEE258 pKa = 3.79HH259 pKa = 6.57ATAAQLTRR267 pKa = 11.84DD268 pKa = 3.63AQKK271 pKa = 11.04RR272 pKa = 11.84LALEE276 pKa = 3.89ADD278 pKa = 4.16GYY280 pKa = 11.48KK281 pKa = 10.53VITVTARR288 pKa = 11.84QIGSQGEE295 pKa = 3.94MRR297 pKa = 11.84KK298 pKa = 9.12IAEE301 pKa = 3.75QSRR304 pKa = 11.84RR305 pKa = 11.84RR306 pKa = 11.84MGTRR310 pKa = 11.84LRR312 pKa = 11.84PQSAAFRR319 pKa = 11.84QQQRR323 pKa = 11.84ALFRR327 pKa = 11.84TGWSLSRR334 pKa = 11.84YY335 pKa = 8.43HH336 pKa = 6.55RR337 pKa = 11.84RR338 pKa = 11.84EE339 pKa = 3.83WLEE342 pKa = 3.82GAPEE346 pKa = 4.03PVAPAPLCPTPVDD359 pKa = 3.45ALTAPGRR366 pKa = 11.84ALRR369 pKa = 11.84CPAVPKK375 pKa = 10.3RR376 pKa = 11.84WPLSDD381 pKa = 4.09AGTFSGLKK389 pKa = 8.94STTLEE394 pKa = 4.2PNSPEE399 pKa = 3.98IEE401 pKa = 4.51GGASPGITPRR411 pKa = 11.84GNTPEE416 pKa = 3.86MM417 pKa = 4.55
MM1 pKa = 7.35KK2 pKa = 10.25AAGGRR7 pKa = 11.84GGRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84TPYY15 pKa = 8.01NTPMRR20 pKa = 11.84IFLYY24 pKa = 10.41GISALSWWLSAPATPGEE41 pKa = 4.26GNRR44 pKa = 11.84VGQNALRR51 pKa = 11.84NCKK54 pKa = 9.22PDD56 pKa = 3.34AAAIQYY62 pKa = 8.16LAHH65 pKa = 7.2CCPQIPRR72 pKa = 11.84PYY74 pKa = 9.35HH75 pKa = 4.31VTVSVRR81 pKa = 11.84MKK83 pKa = 10.12RR84 pKa = 11.84CPDD87 pKa = 2.72GVAPHH92 pKa = 7.15RR93 pKa = 11.84SQFKK97 pKa = 9.78LVGRR101 pKa = 11.84PFCRR105 pKa = 11.84VASGIYY111 pKa = 9.83APCPEE116 pKa = 4.34LCFVQVANNLDD127 pKa = 3.37LHH129 pKa = 6.26EE130 pKa = 4.35LVKK133 pKa = 10.96VGNALCGTFFIDD145 pKa = 3.68PKK147 pKa = 10.86ARR149 pKa = 11.84NGLGKK154 pKa = 10.04RR155 pKa = 11.84RR156 pKa = 11.84PLTSVRR162 pKa = 11.84RR163 pKa = 11.84IGAFIEE169 pKa = 4.3RR170 pKa = 11.84NPGILGAKK178 pKa = 7.85PARR181 pKa = 11.84RR182 pKa = 11.84ALGLMVDD189 pKa = 4.94GAASPPEE196 pKa = 4.25VFLAMALGLPYY207 pKa = 10.56RR208 pKa = 11.84FGGYY212 pKa = 7.66QLPGIAANRR221 pKa = 11.84RR222 pKa = 11.84IKK224 pKa = 10.46PSSKK228 pKa = 10.12ARR230 pKa = 11.84AIAHH234 pKa = 7.08RR235 pKa = 11.84STLVPDD241 pKa = 3.95ILCEE245 pKa = 4.04SSRR248 pKa = 11.84LDD250 pKa = 3.75IEE252 pKa = 4.32YY253 pKa = 10.72DD254 pKa = 3.61SNTEE258 pKa = 3.79HH259 pKa = 6.57ATAAQLTRR267 pKa = 11.84DD268 pKa = 3.63AQKK271 pKa = 11.04RR272 pKa = 11.84LALEE276 pKa = 3.89ADD278 pKa = 4.16GYY280 pKa = 11.48KK281 pKa = 10.53VITVTARR288 pKa = 11.84QIGSQGEE295 pKa = 3.94MRR297 pKa = 11.84KK298 pKa = 9.12IAEE301 pKa = 3.75QSRR304 pKa = 11.84RR305 pKa = 11.84RR306 pKa = 11.84MGTRR310 pKa = 11.84LRR312 pKa = 11.84PQSAAFRR319 pKa = 11.84QQQRR323 pKa = 11.84ALFRR327 pKa = 11.84TGWSLSRR334 pKa = 11.84YY335 pKa = 8.43HH336 pKa = 6.55RR337 pKa = 11.84RR338 pKa = 11.84EE339 pKa = 3.83WLEE342 pKa = 3.82GAPEE346 pKa = 4.03PVAPAPLCPTPVDD359 pKa = 3.45ALTAPGRR366 pKa = 11.84ALRR369 pKa = 11.84CPAVPKK375 pKa = 10.3RR376 pKa = 11.84WPLSDD381 pKa = 4.09AGTFSGLKK389 pKa = 8.94STTLEE394 pKa = 4.2PNSPEE399 pKa = 3.98IEE401 pKa = 4.51GGASPGITPRR411 pKa = 11.84GNTPEE416 pKa = 3.86MM417 pKa = 4.55
Molecular weight: 45.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
846340 |
26 |
24897 |
347.6 |
37.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.98 ± 0.096 | 1.518 ± 0.046 |
6.097 ± 0.116 | 6.724 ± 0.058 |
3.642 ± 0.042 | 8.474 ± 0.048 |
1.671 ± 0.021 | 4.649 ± 0.071 |
3.614 ± 0.055 | 8.96 ± 0.125 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.552 ± 0.049 | 2.851 ± 0.09 |
4.582 ± 0.033 | 2.636 ± 0.026 |
5.992 ± 0.086 | 5.488 ± 0.042 |
5.443 ± 0.117 | 8.05 ± 0.045 |
1.174 ± 0.021 | 2.902 ± 0.076 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
