
Akkermansia glycaniphila
Taxonomy: cellular organisms; Bacteria; PVC group; Verrucomicrobia; Verrucomicrobiae; Verrucomicrobiales; Akkermansiaceae; Akkermansia
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
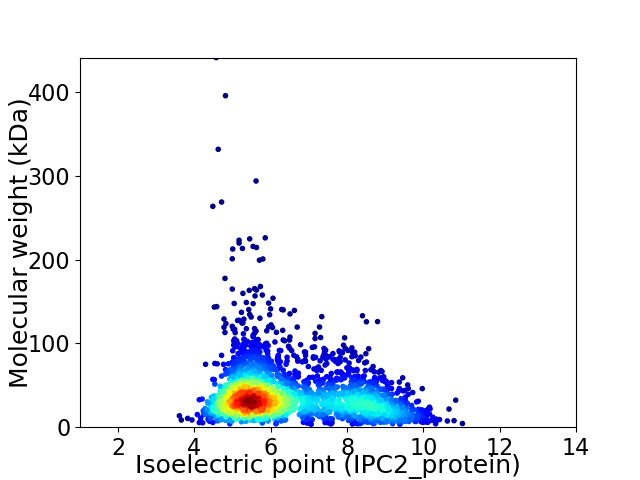
Virtual 2D-PAGE plot for 2530 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1C7PEE3|A0A1C7PEE3_9BACT von willebrand factor type a OS=Akkermansia glycaniphila OX=1679444 GN=PYTT_0121 PE=4 SV=1
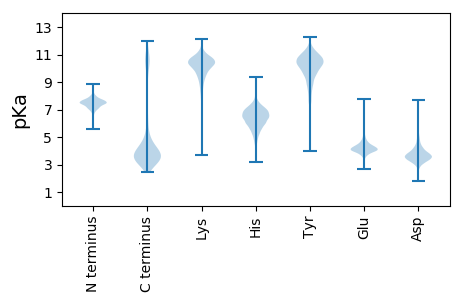
MM1 pKa = 7.57SNNIPDD7 pKa = 3.83NLLYY11 pKa = 10.98SRR13 pKa = 11.84DD14 pKa = 4.03HH15 pKa = 6.36EE16 pKa = 4.49WVDD19 pKa = 3.28VDD21 pKa = 5.38GDD23 pKa = 3.55IATIGISDD31 pKa = 4.01HH32 pKa = 6.44AQAEE36 pKa = 4.33LGDD39 pKa = 4.07VVFVDD44 pKa = 4.81LPEE47 pKa = 5.27PGTSIAANDD56 pKa = 3.99SIAVVEE62 pKa = 4.45SVKK65 pKa = 10.63AASDD69 pKa = 3.79VYY71 pKa = 11.15SPVSGEE77 pKa = 3.64IIEE80 pKa = 4.34VNEE83 pKa = 3.99EE84 pKa = 3.98LGNEE88 pKa = 3.97PGAVNSDD95 pKa = 3.58PYY97 pKa = 11.06GNGWICKK104 pKa = 9.27IRR106 pKa = 11.84LQDD109 pKa = 3.54MSEE112 pKa = 4.14LDD114 pKa = 3.97SLLSPADD121 pKa = 3.49YY122 pKa = 11.47SEE124 pKa = 4.58FCSS127 pKa = 3.96
MM1 pKa = 7.57SNNIPDD7 pKa = 3.83NLLYY11 pKa = 10.98SRR13 pKa = 11.84DD14 pKa = 4.03HH15 pKa = 6.36EE16 pKa = 4.49WVDD19 pKa = 3.28VDD21 pKa = 5.38GDD23 pKa = 3.55IATIGISDD31 pKa = 4.01HH32 pKa = 6.44AQAEE36 pKa = 4.33LGDD39 pKa = 4.07VVFVDD44 pKa = 4.81LPEE47 pKa = 5.27PGTSIAANDD56 pKa = 3.99SIAVVEE62 pKa = 4.45SVKK65 pKa = 10.63AASDD69 pKa = 3.79VYY71 pKa = 11.15SPVSGEE77 pKa = 3.64IIEE80 pKa = 4.34VNEE83 pKa = 3.99EE84 pKa = 3.98LGNEE88 pKa = 3.97PGAVNSDD95 pKa = 3.58PYY97 pKa = 11.06GNGWICKK104 pKa = 9.27IRR106 pKa = 11.84LQDD109 pKa = 3.54MSEE112 pKa = 4.14LDD114 pKa = 3.97SLLSPADD121 pKa = 3.49YY122 pKa = 11.47SEE124 pKa = 4.58FCSS127 pKa = 3.96
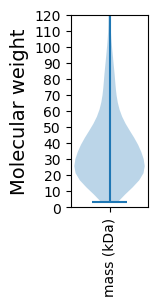
Molecular weight: 13.6 kDa
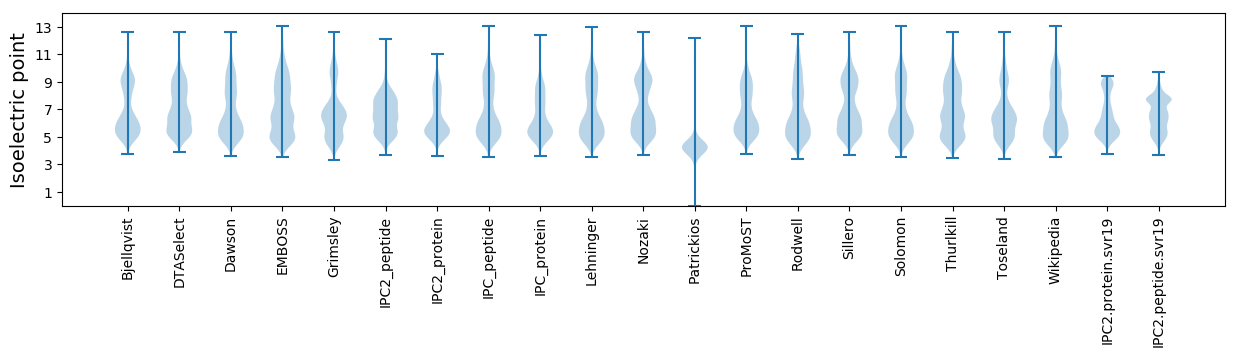
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H6M0L2|A0A1H6M0L2_9BACT 6-phosphofructokinase OS=Akkermansia glycaniphila OX=1679444 GN=PYTT_1977 PE=4 SV=1
MM1 pKa = 7.41LLVEE5 pKa = 4.53KK6 pKa = 9.23TAKK9 pKa = 9.92RR10 pKa = 11.84LKK12 pKa = 10.72SGGLPPVRR20 pKa = 11.84KK21 pKa = 9.87NGLLRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84GFGKK32 pKa = 10.49APARR36 pKa = 11.84LLPPAIRR43 pKa = 11.84MGKK46 pKa = 9.97NIGHH50 pKa = 6.3GAKK53 pKa = 10.28KK54 pKa = 10.22LFEE57 pKa = 4.24TGRR60 pKa = 11.84PPKK63 pKa = 10.34RR64 pKa = 11.84RR65 pKa = 11.84TKK67 pKa = 10.22PILRR71 pKa = 11.84HH72 pKa = 5.43RR73 pKa = 11.84RR74 pKa = 11.84SKK76 pKa = 10.56IRR78 pKa = 11.84TKK80 pKa = 8.6QRR82 pKa = 11.84KK83 pKa = 7.33IAGRR87 pKa = 11.84QIQSRR92 pKa = 11.84RR93 pKa = 11.84RR94 pKa = 11.84IARR97 pKa = 11.84RR98 pKa = 11.84LLARR102 pKa = 11.84HH103 pKa = 5.76RR104 pKa = 11.84QPPQLLPRR112 pKa = 11.84RR113 pKa = 11.84AQRR116 pKa = 11.84THH118 pKa = 6.53LRR120 pKa = 11.84HH121 pKa = 6.19KK122 pKa = 10.11PPEE125 pKa = 3.86PRR127 pKa = 11.84LIRR130 pKa = 11.84RR131 pKa = 11.84SGTRR135 pKa = 11.84LSPPPGSPRR144 pKa = 11.84HH145 pKa = 6.08IPHH148 pKa = 6.32EE149 pKa = 3.74QRR151 pKa = 11.84IRR153 pKa = 11.84IRR155 pKa = 11.84LRR157 pKa = 11.84LRR159 pKa = 11.84MEE161 pKa = 3.92RR162 pKa = 11.84QTTVNRR168 pKa = 11.84RR169 pKa = 11.84PAPLILPRR177 pKa = 11.84PRR179 pKa = 11.84PLRR182 pKa = 11.84LRR184 pKa = 11.84LHH186 pKa = 6.94PLPEE190 pKa = 4.54ILNLQIRR197 pKa = 11.84PLLHH201 pKa = 6.97RR202 pKa = 11.84VQTDD206 pKa = 3.22KK207 pKa = 11.51AHH209 pKa = 6.8PLAFTEE215 pKa = 4.63TAGNTSFTHH224 pKa = 7.4LIDD227 pKa = 3.62LQRR230 pKa = 11.84IQMRR234 pKa = 11.84AQHH237 pKa = 5.69SRR239 pKa = 11.84AGIIAAVIVRR249 pKa = 11.84HH250 pKa = 5.98RR251 pKa = 11.84KK252 pKa = 7.43QAAVKK257 pKa = 9.92HH258 pKa = 5.46LRR260 pKa = 11.84LPITEE265 pKa = 3.94RR266 pKa = 11.84SKK268 pKa = 9.96TIRR271 pKa = 11.84SQNRR275 pKa = 11.84SSS277 pKa = 3.07
MM1 pKa = 7.41LLVEE5 pKa = 4.53KK6 pKa = 9.23TAKK9 pKa = 9.92RR10 pKa = 11.84LKK12 pKa = 10.72SGGLPPVRR20 pKa = 11.84KK21 pKa = 9.87NGLLRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84GFGKK32 pKa = 10.49APARR36 pKa = 11.84LLPPAIRR43 pKa = 11.84MGKK46 pKa = 9.97NIGHH50 pKa = 6.3GAKK53 pKa = 10.28KK54 pKa = 10.22LFEE57 pKa = 4.24TGRR60 pKa = 11.84PPKK63 pKa = 10.34RR64 pKa = 11.84RR65 pKa = 11.84TKK67 pKa = 10.22PILRR71 pKa = 11.84HH72 pKa = 5.43RR73 pKa = 11.84RR74 pKa = 11.84SKK76 pKa = 10.56IRR78 pKa = 11.84TKK80 pKa = 8.6QRR82 pKa = 11.84KK83 pKa = 7.33IAGRR87 pKa = 11.84QIQSRR92 pKa = 11.84RR93 pKa = 11.84RR94 pKa = 11.84IARR97 pKa = 11.84RR98 pKa = 11.84LLARR102 pKa = 11.84HH103 pKa = 5.76RR104 pKa = 11.84QPPQLLPRR112 pKa = 11.84RR113 pKa = 11.84AQRR116 pKa = 11.84THH118 pKa = 6.53LRR120 pKa = 11.84HH121 pKa = 6.19KK122 pKa = 10.11PPEE125 pKa = 3.86PRR127 pKa = 11.84LIRR130 pKa = 11.84RR131 pKa = 11.84SGTRR135 pKa = 11.84LSPPPGSPRR144 pKa = 11.84HH145 pKa = 6.08IPHH148 pKa = 6.32EE149 pKa = 3.74QRR151 pKa = 11.84IRR153 pKa = 11.84IRR155 pKa = 11.84LRR157 pKa = 11.84LRR159 pKa = 11.84MEE161 pKa = 3.92RR162 pKa = 11.84QTTVNRR168 pKa = 11.84RR169 pKa = 11.84PAPLILPRR177 pKa = 11.84PRR179 pKa = 11.84PLRR182 pKa = 11.84LRR184 pKa = 11.84LHH186 pKa = 6.94PLPEE190 pKa = 4.54ILNLQIRR197 pKa = 11.84PLLHH201 pKa = 6.97RR202 pKa = 11.84VQTDD206 pKa = 3.22KK207 pKa = 11.51AHH209 pKa = 6.8PLAFTEE215 pKa = 4.63TAGNTSFTHH224 pKa = 7.4LIDD227 pKa = 3.62LQRR230 pKa = 11.84IQMRR234 pKa = 11.84AQHH237 pKa = 5.69SRR239 pKa = 11.84AGIIAAVIVRR249 pKa = 11.84HH250 pKa = 5.98RR251 pKa = 11.84KK252 pKa = 7.43QAAVKK257 pKa = 9.92HH258 pKa = 5.46LRR260 pKa = 11.84LPITEE265 pKa = 3.94RR266 pKa = 11.84SKK268 pKa = 9.96TIRR271 pKa = 11.84SQNRR275 pKa = 11.84SSS277 pKa = 3.07
Molecular weight: 32.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
925710 |
32 |
4417 |
365.9 |
40.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.942 ± 0.062 | 1.442 ± 0.022 |
5.493 ± 0.031 | 5.989 ± 0.063 |
3.646 ± 0.032 | 7.817 ± 0.088 |
2.201 ± 0.028 | 5.502 ± 0.045 |
4.655 ± 0.053 | 9.615 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.641 ± 0.024 | 3.615 ± 0.058 |
5.096 ± 0.054 | 3.549 ± 0.032 |
5.991 ± 0.067 | 6.126 ± 0.053 |
5.747 ± 0.084 | 6.31 ± 0.057 |
1.512 ± 0.019 | 3.113 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
