
Streptomyces sp. Ag109_O5-1
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; unclassified Streptomyces
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
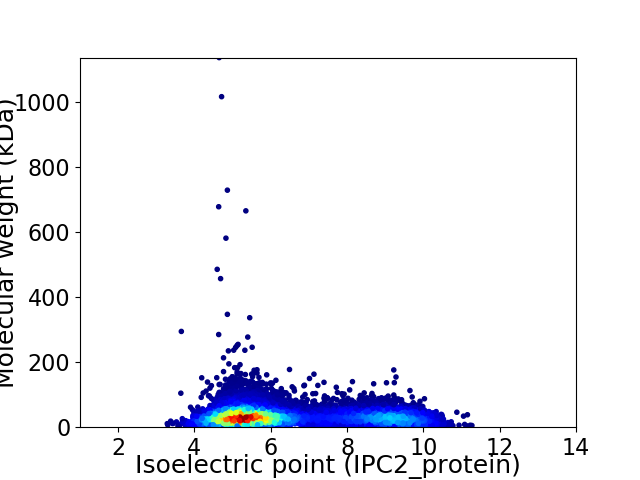
Virtual 2D-PAGE plot for 10263 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
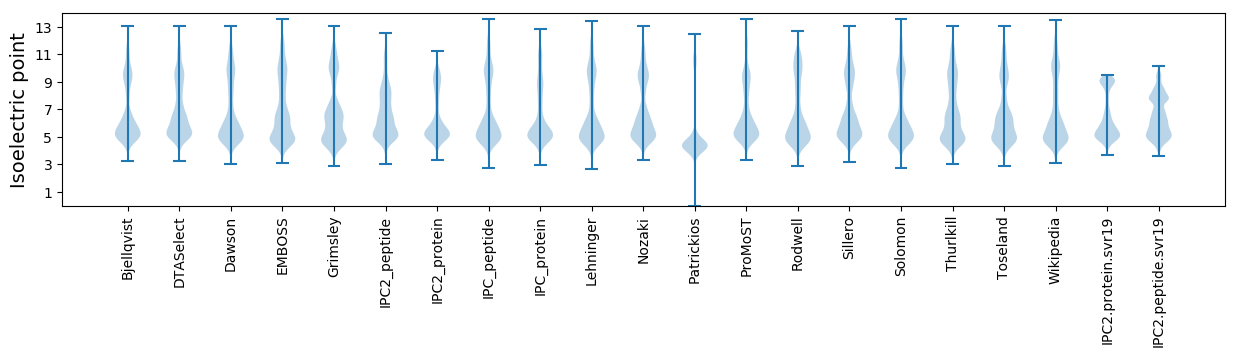
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3N4S336|A0A3N4S336_9ACTN Uncharacterized protein OS=Streptomyces sp. Ag109_O5-1 OX=1938851 GN=EDD90_2844 PE=4 SV=1
MM1 pKa = 7.61AKK3 pKa = 10.23AFQSRR8 pKa = 11.84WRR10 pKa = 11.84QLLGIGRR17 pKa = 11.84NTARR21 pKa = 11.84RR22 pKa = 11.84VAAALLLDD30 pKa = 3.53PTVPVGNFPTGVAINSAGTLAYY52 pKa = 9.78VANEE56 pKa = 4.06GDD58 pKa = 3.98DD59 pKa = 3.86SVSVIDD65 pKa = 3.69TATNAVIATINVGSSPFGVAVTPDD89 pKa = 3.13GAFVYY94 pKa = 8.04VTNVASDD101 pKa = 3.8SVSVIDD107 pKa = 3.7TATNAVTDD115 pKa = 4.13TVSVGISPFGIAINAAGTLAYY136 pKa = 9.49VANVGDD142 pKa = 3.9NTVSVIDD149 pKa = 3.74TSTNTVVTDD158 pKa = 3.6IPVGANPFGVALTPDD173 pKa = 3.19GAFAYY178 pKa = 7.53VTNNASGTVSVVDD191 pKa = 3.42TATNAEE197 pKa = 4.12IDD199 pKa = 4.05TIAVGTSPTGVAITSDD215 pKa = 3.3GVLAYY220 pKa = 8.19VTNNSDD226 pKa = 3.52NTVSAIDD233 pKa = 3.36IATNTVSDD241 pKa = 3.84TFDD244 pKa = 3.15VGSAPTSVAFTPDD257 pKa = 3.07DD258 pKa = 3.48AFAYY262 pKa = 7.0VTNNASDD269 pKa = 3.77TVSVVITATNDD280 pKa = 2.96VTFDD284 pKa = 3.47IPVGSGPFGVAITPDD299 pKa = 3.63GEE301 pKa = 4.21HH302 pKa = 7.3AYY304 pKa = 8.65VTNNASDD311 pKa = 3.97TVSVTDD317 pKa = 3.51TVPFPTTTTLTSAPDD332 pKa = 3.55PSAFGEE338 pKa = 4.27AKK340 pKa = 10.54VLTATVTSSTGTPDD354 pKa = 3.14GTVTFLDD361 pKa = 4.29GATPLGTVTLAGGVATLTTSSLAVGSHH388 pKa = 6.26SLTADD393 pKa = 3.2YY394 pKa = 10.95AGNVNFSVSTSAADD408 pKa = 3.3IQTVNAADD416 pKa = 3.69TATTLTSSPDD426 pKa = 3.23PSVFGEE432 pKa = 4.14AKK434 pKa = 10.48VLTATVTAVAPGSGTPTGSVSFLDD458 pKa = 3.99GATVIGIGTLDD469 pKa = 3.67GGGVATFSTSSLSAGTHH486 pKa = 5.8SLTAVYY492 pKa = 10.44AGDD495 pKa = 3.71GSFNGSTSPVDD506 pKa = 3.76TQTVTAANTTTALTSSPDD524 pKa = 3.37PSVFGQAKK532 pKa = 9.82VLTATVTAVAPGAGVPTGTVTFFDD556 pKa = 4.19GMTSLGTGTLDD567 pKa = 3.36GSGVATLSVSNLSVGAHH584 pKa = 6.22SLTAGYY590 pKa = 10.69AGTADD595 pKa = 4.09YY596 pKa = 11.17NSSTSAVDD604 pKa = 3.57SQTVTAANTTTALTSSPDD622 pKa = 3.37PSVFGQAKK630 pKa = 9.71VLTATVSAVAPGAGVPTGTVTFFDD654 pKa = 4.19GMTSLGTGTLNGSGVATLSVSNLSVGAHH682 pKa = 6.22SLTAGYY688 pKa = 10.69AGTADD693 pKa = 4.09YY694 pKa = 11.17NSSTSAVDD702 pKa = 3.57SQTVTAANTTTALTSSPDD720 pKa = 3.37PSVFGQAKK728 pKa = 9.71VLTATVSAVAPGAGVPTGTVTFFDD752 pKa = 4.19GMTSLGTGTLNGSGVATLSVSNLSVGAHH780 pKa = 6.22SLTAGYY786 pKa = 10.69AGTADD791 pKa = 4.09YY792 pKa = 11.17NSSTSAVDD800 pKa = 3.57SQTVTAANTTTALTSSPDD818 pKa = 3.38PSVFGQGKK826 pKa = 9.01VLTATVSAVAPGAGVPTGTVTFFDD850 pKa = 4.19GMTSLGTGILVGSGVATLSVSNLSVGSHH878 pKa = 6.86ALTAAYY884 pKa = 10.05GGDD887 pKa = 3.13TSYY890 pKa = 11.55NGSTSPIDD898 pKa = 3.69TQTVNAADD906 pKa = 3.74TATALTSSPDD916 pKa = 3.29PSVFGEE922 pKa = 4.14AKK924 pKa = 10.44VLTATVSAVAPGAGVPTGTVTFFDD948 pKa = 4.19GMTSLGTGILNGSGVATLSVSNLSVGSHH976 pKa = 6.86ALTAAYY982 pKa = 10.05GGDD985 pKa = 3.13TSYY988 pKa = 11.55NGSTSPIDD996 pKa = 3.69TQTVNAADD1004 pKa = 3.74TTTALTSSPDD1014 pKa = 3.29PSVFGEE1020 pKa = 4.45TKK1022 pKa = 10.61VLTATVTPVAPGTGVPTGTVTFFDD1046 pKa = 4.19GMTSLGTGTLDD1057 pKa = 3.36GSGVATLSVSNLSVGTHH1074 pKa = 7.12PLTAAYY1080 pKa = 10.06GGDD1083 pKa = 3.13TSYY1086 pKa = 11.52NGSTSPVDD1094 pKa = 3.66TQTVNAADD1102 pKa = 3.74TATALTSSPDD1112 pKa = 3.29PSVFGEE1118 pKa = 4.14AKK1120 pKa = 10.48VLTATVTAVAPGSGTPTGSVSFLDD1144 pKa = 3.99GATVIGIGTLDD1155 pKa = 3.67GGGVATFTTSSLSAGTHH1172 pKa = 5.8SLTAVYY1178 pKa = 10.44AGDD1181 pKa = 3.71GSFNGSTSPVDD1192 pKa = 3.66TQTVNAANTTTALTSSPDD1210 pKa = 3.29PSVFGEE1216 pKa = 4.14AKK1218 pKa = 10.44VLTATVSAVAPGAGVPTGTVTFFDD1242 pKa = 4.19GMTSLGTGTLDD1253 pKa = 3.36GSGVATLSVSNLSVGSHH1270 pKa = 6.86ALTAAYY1276 pKa = 10.05GGDD1279 pKa = 3.13TSYY1282 pKa = 11.55NGSTSPIDD1290 pKa = 3.69TQTVNAADD1298 pKa = 3.74TATALTSSPDD1308 pKa = 3.29PSVFGEE1314 pKa = 4.14AKK1316 pKa = 10.44VLTATVSAVAPGAGVPTGTVTFFDD1340 pKa = 4.19GMTSLGTGILDD1351 pKa = 3.69GSGVATLSVSNLSVGTHH1368 pKa = 7.12PLTAAYY1374 pKa = 10.06GGDD1377 pKa = 3.13TSYY1380 pKa = 11.55NGSTSPIDD1388 pKa = 3.69TQTVNAANTTTALTSSPDD1406 pKa = 3.29PSVFGEE1412 pKa = 4.45TKK1414 pKa = 10.61VLTATVTPVAPGTGVPTGTVTFFDD1438 pKa = 4.19GMTSLGTGTLDD1449 pKa = 3.36GSGVATLSVSNLSVGTHH1466 pKa = 7.12PLTAAYY1472 pKa = 10.06GGDD1475 pKa = 3.13TSYY1478 pKa = 11.52NGSTSPVDD1486 pKa = 3.66TQTVNAADD1494 pKa = 3.74TATALTSSPDD1504 pKa = 3.29PSVFGEE1510 pKa = 4.14AKK1512 pKa = 10.48VLTATVTAVAPGSGTPTGSVSFLDD1536 pKa = 3.99GATVIGIGTLDD1547 pKa = 3.67GGGVATFTTSSLSAGTHH1564 pKa = 5.8SLTAVYY1570 pKa = 10.44AGDD1573 pKa = 3.71GSFNGSTSPVDD1584 pKa = 3.66TQTVNAANTTTALTSSPDD1602 pKa = 3.29PSVFGEE1608 pKa = 4.14AKK1610 pKa = 10.44VLTATVSAVAPGAGVPTGTVTFFDD1634 pKa = 4.19GMTSLGTGTLDD1645 pKa = 3.36GSGVATLSVSNLSVGTHH1662 pKa = 7.12PLTAAYY1668 pKa = 10.06GGDD1671 pKa = 3.13TSYY1674 pKa = 11.58NGSTSPTDD1682 pKa = 3.51TQTVNAADD1690 pKa = 3.74TTTALTSSPDD1700 pKa = 3.36PSVFGQTKK1708 pKa = 9.87VLTATVSAVAPGAGVPTGTVTFFDD1732 pKa = 4.19GMTSLGTGTLDD1743 pKa = 3.4GTGVATLSVSNLSVGTHH1760 pKa = 7.05PLTATYY1766 pKa = 10.72AGDD1769 pKa = 3.34TSYY1772 pKa = 11.5NGSTSPIDD1780 pKa = 3.69TQTVNAADD1788 pKa = 3.74TATTLTSSPDD1798 pKa = 3.31PSVFGQAKK1806 pKa = 9.71VLTATVSPVAPGGGIPTGTVTFFDD1830 pKa = 4.19GMTSLGTGTLDD1841 pKa = 3.4GTGVATLSVSNLSVGTHH1858 pKa = 7.09PLTATYY1864 pKa = 10.67GGDD1867 pKa = 3.25SSFNGSTSPADD1878 pKa = 3.71TQTVNAANTTTALTSSPDD1896 pKa = 3.38PSVFGQGKK1904 pKa = 9.01VLTATVSPVAPGGGIPTGTVTFFDD1928 pKa = 4.19GMTSLGTGTLDD1939 pKa = 3.36GSGVATLTVSNLSVGSHH1956 pKa = 6.8ALTATYY1962 pKa = 10.6GGDD1965 pKa = 3.25SSFNGSTSPADD1976 pKa = 3.71TQTVNAANTTTTLTSSPDD1994 pKa = 3.23PSVFGEE2000 pKa = 4.14AKK2002 pKa = 10.48VLTATVTAVAPGSGTPTGSVSFLDD2026 pKa = 3.99GATVIGIGTLDD2037 pKa = 3.67GGGVATFTTSTLGAGTHH2054 pKa = 6.08SLTAVYY2060 pKa = 10.44AGDD2063 pKa = 3.71GSFNGSTSPVDD2074 pKa = 3.66TQTVNAANTTTALTSSPDD2092 pKa = 3.38PSVFGQGKK2100 pKa = 9.01VLTATVSPVAPGGGIPTGTVTFFDD2124 pKa = 4.19GMTSLGTGTLNGSGVATLTVSNLSVGSHH2152 pKa = 6.18SLTATYY2158 pKa = 10.66GGDD2161 pKa = 3.25SSFNGSTSPADD2172 pKa = 3.71TQTVNAADD2180 pKa = 3.74TTTALTSSPDD2190 pKa = 3.36PSVFGQTKK2198 pKa = 9.87VLTATVSAVAPGAGVPTGTVTFFDD2222 pKa = 4.19GMTSLGTGTLDD2233 pKa = 3.4GTGVATLSVSNLSVGTHH2250 pKa = 7.09PLTATYY2256 pKa = 10.63GGDD2259 pKa = 3.01TSYY2262 pKa = 11.59NGSTSPADD2270 pKa = 3.58TQTVNTANTTATLTSSPDD2288 pKa = 3.29PSVFGQTKK2296 pKa = 10.04VLTATVTPVAPGTGVPTGTVTFFDD2320 pKa = 4.19GMTSLGTGTLDD2331 pKa = 3.36GSGVATLSVSNLSVGTHH2348 pKa = 7.12PLTAAYY2354 pKa = 10.11GGDD2357 pKa = 3.49TSHH2360 pKa = 6.95NGSTSPIDD2368 pKa = 3.69TQTVNAANTTTALTSSPDD2386 pKa = 3.36PSVFGQTKK2394 pKa = 10.04VLTATVTPVAPGTGVPTGTVTFFDD2418 pKa = 4.19GMTSLGTGTLNGSGVATLSVSNLSVGTHH2446 pKa = 7.12PLTAAYY2452 pKa = 10.11GGDD2455 pKa = 3.49TSHH2458 pKa = 6.94NGSTSPVDD2466 pKa = 3.4TQTVNKK2472 pKa = 10.28ANTTTAVSSSPNPSVFGQSVNLTATVAVVAPGGGVPGGTVTFFVGGIPQTPAVPLNGSGVATLSISTLSVGTRR2545 pKa = 11.84SVRR2548 pKa = 11.84ASYY2551 pKa = 10.65NGSAGYY2557 pKa = 7.79NTSTSPTISQVVSKK2571 pKa = 11.02ANTSTTLTSSPDD2583 pKa = 3.31PSVFGQTKK2591 pKa = 9.87VLTATVSPVAPGGGTPIGTVSFFDD2615 pKa = 3.94GMTLLGTGTLSGGVATLSTSALALGSHH2642 pKa = 6.51SLTATYY2648 pKa = 10.36GGSANFNGSTSPVDD2662 pKa = 3.62TQTVNPANTTTVLTSSPDD2680 pKa = 3.51PSVLGQAKK2688 pKa = 10.02VLTATVSATAPGAGTPSGTVTFFDD2712 pKa = 4.26GMTSLGTGALDD2723 pKa = 3.79GSGVATLSVSNLSVGSHH2740 pKa = 5.89TLTATYY2746 pKa = 10.29GGSANFNGSTSAVDD2760 pKa = 3.58TQTVNPANTMTALTSSPDD2778 pKa = 3.49PSVTGQAKK2786 pKa = 10.08VLTATVTAVAPGTGTPSGSVTFFDD2810 pKa = 5.01GMTSLGTGTLDD2821 pKa = 3.36GSGVATLSVSNLSVGNHH2838 pKa = 6.07SLTAAYY2844 pKa = 9.67GGSGSYY2850 pKa = 10.26NGSTSSADD2858 pKa = 3.31TQTVNKK2864 pKa = 10.27ANTTTAVSSSPNPSVFGQSVNLTATVAVVAPGGGVPGGTVTFFIGGIPQTPAVPLNGSGVATLPISTLSVGTRR2937 pKa = 11.84SVRR2940 pKa = 11.84ASYY2943 pKa = 10.65NGSAGYY2949 pKa = 7.79NTSTSPTISQVVSKK2963 pKa = 11.02ANTSTTLVSAPNPSMSGQSVTLTATVAPVAPGGGVPTGSVSFLDD3007 pKa = 4.3GMTLLGTGTLSGGVATFSTSSLSVGSHH3034 pKa = 6.1SLTAAYY3040 pKa = 9.84AGSGSYY3046 pKa = 10.23NGSTSPAVTQTVSS3059 pKa = 2.63
MM1 pKa = 7.61AKK3 pKa = 10.23AFQSRR8 pKa = 11.84WRR10 pKa = 11.84QLLGIGRR17 pKa = 11.84NTARR21 pKa = 11.84RR22 pKa = 11.84VAAALLLDD30 pKa = 3.53PTVPVGNFPTGVAINSAGTLAYY52 pKa = 9.78VANEE56 pKa = 4.06GDD58 pKa = 3.98DD59 pKa = 3.86SVSVIDD65 pKa = 3.69TATNAVIATINVGSSPFGVAVTPDD89 pKa = 3.13GAFVYY94 pKa = 8.04VTNVASDD101 pKa = 3.8SVSVIDD107 pKa = 3.7TATNAVTDD115 pKa = 4.13TVSVGISPFGIAINAAGTLAYY136 pKa = 9.49VANVGDD142 pKa = 3.9NTVSVIDD149 pKa = 3.74TSTNTVVTDD158 pKa = 3.6IPVGANPFGVALTPDD173 pKa = 3.19GAFAYY178 pKa = 7.53VTNNASGTVSVVDD191 pKa = 3.42TATNAEE197 pKa = 4.12IDD199 pKa = 4.05TIAVGTSPTGVAITSDD215 pKa = 3.3GVLAYY220 pKa = 8.19VTNNSDD226 pKa = 3.52NTVSAIDD233 pKa = 3.36IATNTVSDD241 pKa = 3.84TFDD244 pKa = 3.15VGSAPTSVAFTPDD257 pKa = 3.07DD258 pKa = 3.48AFAYY262 pKa = 7.0VTNNASDD269 pKa = 3.77TVSVVITATNDD280 pKa = 2.96VTFDD284 pKa = 3.47IPVGSGPFGVAITPDD299 pKa = 3.63GEE301 pKa = 4.21HH302 pKa = 7.3AYY304 pKa = 8.65VTNNASDD311 pKa = 3.97TVSVTDD317 pKa = 3.51TVPFPTTTTLTSAPDD332 pKa = 3.55PSAFGEE338 pKa = 4.27AKK340 pKa = 10.54VLTATVTSSTGTPDD354 pKa = 3.14GTVTFLDD361 pKa = 4.29GATPLGTVTLAGGVATLTTSSLAVGSHH388 pKa = 6.26SLTADD393 pKa = 3.2YY394 pKa = 10.95AGNVNFSVSTSAADD408 pKa = 3.3IQTVNAADD416 pKa = 3.69TATTLTSSPDD426 pKa = 3.23PSVFGEE432 pKa = 4.14AKK434 pKa = 10.48VLTATVTAVAPGSGTPTGSVSFLDD458 pKa = 3.99GATVIGIGTLDD469 pKa = 3.67GGGVATFSTSSLSAGTHH486 pKa = 5.8SLTAVYY492 pKa = 10.44AGDD495 pKa = 3.71GSFNGSTSPVDD506 pKa = 3.76TQTVTAANTTTALTSSPDD524 pKa = 3.37PSVFGQAKK532 pKa = 9.82VLTATVTAVAPGAGVPTGTVTFFDD556 pKa = 4.19GMTSLGTGTLDD567 pKa = 3.36GSGVATLSVSNLSVGAHH584 pKa = 6.22SLTAGYY590 pKa = 10.69AGTADD595 pKa = 4.09YY596 pKa = 11.17NSSTSAVDD604 pKa = 3.57SQTVTAANTTTALTSSPDD622 pKa = 3.37PSVFGQAKK630 pKa = 9.71VLTATVSAVAPGAGVPTGTVTFFDD654 pKa = 4.19GMTSLGTGTLNGSGVATLSVSNLSVGAHH682 pKa = 6.22SLTAGYY688 pKa = 10.69AGTADD693 pKa = 4.09YY694 pKa = 11.17NSSTSAVDD702 pKa = 3.57SQTVTAANTTTALTSSPDD720 pKa = 3.37PSVFGQAKK728 pKa = 9.71VLTATVSAVAPGAGVPTGTVTFFDD752 pKa = 4.19GMTSLGTGTLNGSGVATLSVSNLSVGAHH780 pKa = 6.22SLTAGYY786 pKa = 10.69AGTADD791 pKa = 4.09YY792 pKa = 11.17NSSTSAVDD800 pKa = 3.57SQTVTAANTTTALTSSPDD818 pKa = 3.38PSVFGQGKK826 pKa = 9.01VLTATVSAVAPGAGVPTGTVTFFDD850 pKa = 4.19GMTSLGTGILVGSGVATLSVSNLSVGSHH878 pKa = 6.86ALTAAYY884 pKa = 10.05GGDD887 pKa = 3.13TSYY890 pKa = 11.55NGSTSPIDD898 pKa = 3.69TQTVNAADD906 pKa = 3.74TATALTSSPDD916 pKa = 3.29PSVFGEE922 pKa = 4.14AKK924 pKa = 10.44VLTATVSAVAPGAGVPTGTVTFFDD948 pKa = 4.19GMTSLGTGILNGSGVATLSVSNLSVGSHH976 pKa = 6.86ALTAAYY982 pKa = 10.05GGDD985 pKa = 3.13TSYY988 pKa = 11.55NGSTSPIDD996 pKa = 3.69TQTVNAADD1004 pKa = 3.74TTTALTSSPDD1014 pKa = 3.29PSVFGEE1020 pKa = 4.45TKK1022 pKa = 10.61VLTATVTPVAPGTGVPTGTVTFFDD1046 pKa = 4.19GMTSLGTGTLDD1057 pKa = 3.36GSGVATLSVSNLSVGTHH1074 pKa = 7.12PLTAAYY1080 pKa = 10.06GGDD1083 pKa = 3.13TSYY1086 pKa = 11.52NGSTSPVDD1094 pKa = 3.66TQTVNAADD1102 pKa = 3.74TATALTSSPDD1112 pKa = 3.29PSVFGEE1118 pKa = 4.14AKK1120 pKa = 10.48VLTATVTAVAPGSGTPTGSVSFLDD1144 pKa = 3.99GATVIGIGTLDD1155 pKa = 3.67GGGVATFTTSSLSAGTHH1172 pKa = 5.8SLTAVYY1178 pKa = 10.44AGDD1181 pKa = 3.71GSFNGSTSPVDD1192 pKa = 3.66TQTVNAANTTTALTSSPDD1210 pKa = 3.29PSVFGEE1216 pKa = 4.14AKK1218 pKa = 10.44VLTATVSAVAPGAGVPTGTVTFFDD1242 pKa = 4.19GMTSLGTGTLDD1253 pKa = 3.36GSGVATLSVSNLSVGSHH1270 pKa = 6.86ALTAAYY1276 pKa = 10.05GGDD1279 pKa = 3.13TSYY1282 pKa = 11.55NGSTSPIDD1290 pKa = 3.69TQTVNAADD1298 pKa = 3.74TATALTSSPDD1308 pKa = 3.29PSVFGEE1314 pKa = 4.14AKK1316 pKa = 10.44VLTATVSAVAPGAGVPTGTVTFFDD1340 pKa = 4.19GMTSLGTGILDD1351 pKa = 3.69GSGVATLSVSNLSVGTHH1368 pKa = 7.12PLTAAYY1374 pKa = 10.06GGDD1377 pKa = 3.13TSYY1380 pKa = 11.55NGSTSPIDD1388 pKa = 3.69TQTVNAANTTTALTSSPDD1406 pKa = 3.29PSVFGEE1412 pKa = 4.45TKK1414 pKa = 10.61VLTATVTPVAPGTGVPTGTVTFFDD1438 pKa = 4.19GMTSLGTGTLDD1449 pKa = 3.36GSGVATLSVSNLSVGTHH1466 pKa = 7.12PLTAAYY1472 pKa = 10.06GGDD1475 pKa = 3.13TSYY1478 pKa = 11.52NGSTSPVDD1486 pKa = 3.66TQTVNAADD1494 pKa = 3.74TATALTSSPDD1504 pKa = 3.29PSVFGEE1510 pKa = 4.14AKK1512 pKa = 10.48VLTATVTAVAPGSGTPTGSVSFLDD1536 pKa = 3.99GATVIGIGTLDD1547 pKa = 3.67GGGVATFTTSSLSAGTHH1564 pKa = 5.8SLTAVYY1570 pKa = 10.44AGDD1573 pKa = 3.71GSFNGSTSPVDD1584 pKa = 3.66TQTVNAANTTTALTSSPDD1602 pKa = 3.29PSVFGEE1608 pKa = 4.14AKK1610 pKa = 10.44VLTATVSAVAPGAGVPTGTVTFFDD1634 pKa = 4.19GMTSLGTGTLDD1645 pKa = 3.36GSGVATLSVSNLSVGTHH1662 pKa = 7.12PLTAAYY1668 pKa = 10.06GGDD1671 pKa = 3.13TSYY1674 pKa = 11.58NGSTSPTDD1682 pKa = 3.51TQTVNAADD1690 pKa = 3.74TTTALTSSPDD1700 pKa = 3.36PSVFGQTKK1708 pKa = 9.87VLTATVSAVAPGAGVPTGTVTFFDD1732 pKa = 4.19GMTSLGTGTLDD1743 pKa = 3.4GTGVATLSVSNLSVGTHH1760 pKa = 7.05PLTATYY1766 pKa = 10.72AGDD1769 pKa = 3.34TSYY1772 pKa = 11.5NGSTSPIDD1780 pKa = 3.69TQTVNAADD1788 pKa = 3.74TATTLTSSPDD1798 pKa = 3.31PSVFGQAKK1806 pKa = 9.71VLTATVSPVAPGGGIPTGTVTFFDD1830 pKa = 4.19GMTSLGTGTLDD1841 pKa = 3.4GTGVATLSVSNLSVGTHH1858 pKa = 7.09PLTATYY1864 pKa = 10.67GGDD1867 pKa = 3.25SSFNGSTSPADD1878 pKa = 3.71TQTVNAANTTTALTSSPDD1896 pKa = 3.38PSVFGQGKK1904 pKa = 9.01VLTATVSPVAPGGGIPTGTVTFFDD1928 pKa = 4.19GMTSLGTGTLDD1939 pKa = 3.36GSGVATLTVSNLSVGSHH1956 pKa = 6.8ALTATYY1962 pKa = 10.6GGDD1965 pKa = 3.25SSFNGSTSPADD1976 pKa = 3.71TQTVNAANTTTTLTSSPDD1994 pKa = 3.23PSVFGEE2000 pKa = 4.14AKK2002 pKa = 10.48VLTATVTAVAPGSGTPTGSVSFLDD2026 pKa = 3.99GATVIGIGTLDD2037 pKa = 3.67GGGVATFTTSTLGAGTHH2054 pKa = 6.08SLTAVYY2060 pKa = 10.44AGDD2063 pKa = 3.71GSFNGSTSPVDD2074 pKa = 3.66TQTVNAANTTTALTSSPDD2092 pKa = 3.38PSVFGQGKK2100 pKa = 9.01VLTATVSPVAPGGGIPTGTVTFFDD2124 pKa = 4.19GMTSLGTGTLNGSGVATLTVSNLSVGSHH2152 pKa = 6.18SLTATYY2158 pKa = 10.66GGDD2161 pKa = 3.25SSFNGSTSPADD2172 pKa = 3.71TQTVNAADD2180 pKa = 3.74TTTALTSSPDD2190 pKa = 3.36PSVFGQTKK2198 pKa = 9.87VLTATVSAVAPGAGVPTGTVTFFDD2222 pKa = 4.19GMTSLGTGTLDD2233 pKa = 3.4GTGVATLSVSNLSVGTHH2250 pKa = 7.09PLTATYY2256 pKa = 10.63GGDD2259 pKa = 3.01TSYY2262 pKa = 11.59NGSTSPADD2270 pKa = 3.58TQTVNTANTTATLTSSPDD2288 pKa = 3.29PSVFGQTKK2296 pKa = 10.04VLTATVTPVAPGTGVPTGTVTFFDD2320 pKa = 4.19GMTSLGTGTLDD2331 pKa = 3.36GSGVATLSVSNLSVGTHH2348 pKa = 7.12PLTAAYY2354 pKa = 10.11GGDD2357 pKa = 3.49TSHH2360 pKa = 6.95NGSTSPIDD2368 pKa = 3.69TQTVNAANTTTALTSSPDD2386 pKa = 3.36PSVFGQTKK2394 pKa = 10.04VLTATVTPVAPGTGVPTGTVTFFDD2418 pKa = 4.19GMTSLGTGTLNGSGVATLSVSNLSVGTHH2446 pKa = 7.12PLTAAYY2452 pKa = 10.11GGDD2455 pKa = 3.49TSHH2458 pKa = 6.94NGSTSPVDD2466 pKa = 3.4TQTVNKK2472 pKa = 10.28ANTTTAVSSSPNPSVFGQSVNLTATVAVVAPGGGVPGGTVTFFVGGIPQTPAVPLNGSGVATLSISTLSVGTRR2545 pKa = 11.84SVRR2548 pKa = 11.84ASYY2551 pKa = 10.65NGSAGYY2557 pKa = 7.79NTSTSPTISQVVSKK2571 pKa = 11.02ANTSTTLTSSPDD2583 pKa = 3.31PSVFGQTKK2591 pKa = 9.87VLTATVSPVAPGGGTPIGTVSFFDD2615 pKa = 3.94GMTLLGTGTLSGGVATLSTSALALGSHH2642 pKa = 6.51SLTATYY2648 pKa = 10.36GGSANFNGSTSPVDD2662 pKa = 3.62TQTVNPANTTTVLTSSPDD2680 pKa = 3.51PSVLGQAKK2688 pKa = 10.02VLTATVSATAPGAGTPSGTVTFFDD2712 pKa = 4.26GMTSLGTGALDD2723 pKa = 3.79GSGVATLSVSNLSVGSHH2740 pKa = 5.89TLTATYY2746 pKa = 10.29GGSANFNGSTSAVDD2760 pKa = 3.58TQTVNPANTMTALTSSPDD2778 pKa = 3.49PSVTGQAKK2786 pKa = 10.08VLTATVTAVAPGTGTPSGSVTFFDD2810 pKa = 5.01GMTSLGTGTLDD2821 pKa = 3.36GSGVATLSVSNLSVGNHH2838 pKa = 6.07SLTAAYY2844 pKa = 9.67GGSGSYY2850 pKa = 10.26NGSTSSADD2858 pKa = 3.31TQTVNKK2864 pKa = 10.27ANTTTAVSSSPNPSVFGQSVNLTATVAVVAPGGGVPGGTVTFFIGGIPQTPAVPLNGSGVATLPISTLSVGTRR2937 pKa = 11.84SVRR2940 pKa = 11.84ASYY2943 pKa = 10.65NGSAGYY2949 pKa = 7.79NTSTSPTISQVVSKK2963 pKa = 11.02ANTSTTLVSAPNPSMSGQSVTLTATVAPVAPGGGVPTGSVSFLDD3007 pKa = 4.3GMTLLGTGTLSGGVATFSTSSLSVGSHH3034 pKa = 6.1SLTAAYY3040 pKa = 9.84AGSGSYY3046 pKa = 10.23NGSTSPAVTQTVSS3059 pKa = 2.63
Molecular weight: 294.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3N4SG66|A0A3N4SG66_9ACTN Uncharacterized protein OS=Streptomyces sp. Ag109_O5-1 OX=1938851 GN=EDD90_5732 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILANRR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.46GRR40 pKa = 11.84ASLSAA45 pKa = 3.83
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILANRR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.46GRR40 pKa = 11.84ASLSAA45 pKa = 3.83
Molecular weight: 5.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3383640 |
29 |
10866 |
329.7 |
35.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.353 ± 0.035 | 0.805 ± 0.007 |
5.968 ± 0.019 | 5.445 ± 0.026 |
2.754 ± 0.014 | 9.323 ± 0.024 |
2.376 ± 0.015 | 3.201 ± 0.017 |
2.181 ± 0.023 | 10.246 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.702 ± 0.01 | 1.893 ± 0.013 |
6.032 ± 0.021 | 2.895 ± 0.017 |
7.889 ± 0.031 | 5.271 ± 0.022 |
6.434 ± 0.028 | 8.459 ± 0.043 |
1.582 ± 0.01 | 2.191 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
