
Lacibacter luteus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Chitinophagia; Chitinophagales; Chitinophagaceae; Lacibacter
Average proteome isoelectric point is 7.02
Get precalculated fractions of proteins
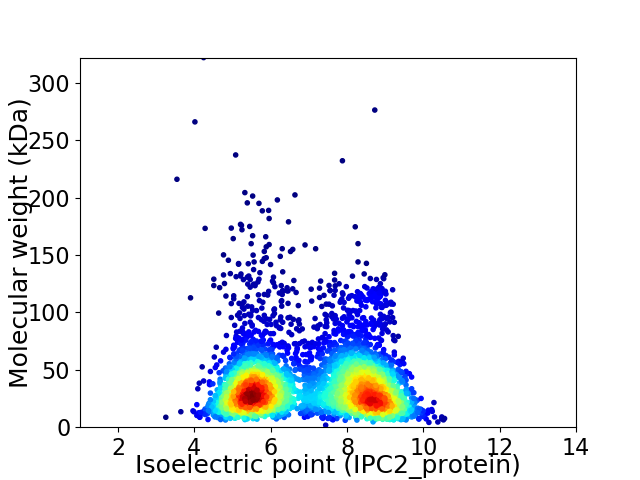
Virtual 2D-PAGE plot for 4180 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q1CNH8|A0A4Q1CNH8_9BACT Beta-glucosidase OS=Lacibacter luteus OX=2508719 GN=ESA94_04415 PE=4 SV=1
MM1 pKa = 7.29KK2 pKa = 10.46QFLPYY7 pKa = 9.97KK8 pKa = 8.45RR9 pKa = 11.84NCMGAAIKK17 pKa = 10.24NLHH20 pKa = 6.56LLFTLSIALFVSTNVANAQCTCTGNIVVNPSFEE53 pKa = 4.39SGTTGWSWSGGSFNAGTGAIACGSKK78 pKa = 10.49SGDD81 pKa = 3.42FQISNTSSNWVSQTIGTDD99 pKa = 3.72LAPGTKK105 pKa = 9.47INASVYY111 pKa = 10.77AGTHH115 pKa = 5.07NNSYY119 pKa = 8.75YY120 pKa = 10.35HH121 pKa = 5.9QVAIDD126 pKa = 3.92FFDD129 pKa = 5.82ANWNWISNSKK139 pKa = 10.38VEE141 pKa = 4.21VNKK144 pKa = 10.1VLSTSPVGPQLYY156 pKa = 8.27TWSGTVPVGTKK167 pKa = 7.39YY168 pKa = 9.61TNVSFSGNGDD178 pKa = 3.84WIKK181 pKa = 10.2TDD183 pKa = 3.21QWCVTLTPPAGGSIGDD199 pKa = 4.23RR200 pKa = 11.84VWLDD204 pKa = 3.06ADD206 pKa = 4.07GDD208 pKa = 4.27GLQDD212 pKa = 3.4ASEE215 pKa = 4.26TSGIAGVTVQLKK227 pKa = 10.25NSLGSVIATTTTNASGNYY245 pKa = 9.76LFTGLAAGNYY255 pKa = 6.93TVVFPTSISGAVVTPQNVGLDD276 pKa = 3.97DD277 pKa = 4.96NVDD280 pKa = 3.47SDD282 pKa = 4.49ASQTTGATSTITLATNQNITNVDD305 pKa = 3.06AGYY308 pKa = 10.83CPTTLSLGNRR318 pKa = 11.84VWYY321 pKa = 7.73DD322 pKa = 3.48TNNDD326 pKa = 4.14GINSNEE332 pKa = 3.8NGIPNLTVYY341 pKa = 10.55LYY343 pKa = 11.09KK344 pKa = 10.48DD345 pKa = 3.74DD346 pKa = 5.55NNDD349 pKa = 3.19NVADD353 pKa = 3.93AAAIATAVTDD363 pKa = 3.82ANGYY367 pKa = 10.47YY368 pKa = 9.01MFSNLAPGNYY378 pKa = 8.66IVGAVTPAGYY388 pKa = 9.23MSSLVNGGDD397 pKa = 3.46PDD399 pKa = 5.01DD400 pKa = 5.17NNNSDD405 pKa = 4.55DD406 pKa = 3.97NGEE409 pKa = 4.24VTVGAEE415 pKa = 4.05TRR417 pKa = 11.84GLAITLVAGTEE428 pKa = 3.99PLVDD432 pKa = 3.89GNTNSNSNITYY443 pKa = 10.52DD444 pKa = 4.2FGFLPDD450 pKa = 4.37CNCTSSSSNLLINGSFEE467 pKa = 4.18NGTTGWSASGGSVSSGTGYY486 pKa = 9.57VACGAANGFNSWSSGTSRR504 pKa = 11.84VWQDD508 pKa = 2.87VAVAAGSTVVFKK520 pKa = 11.02GFAGVHH526 pKa = 4.68TAGLTCSPKK535 pKa = 10.61LSLIFYY541 pKa = 9.14NSSNTVISQNDD552 pKa = 3.44VVVTRR557 pKa = 11.84DD558 pKa = 2.69VDD560 pKa = 3.52KK561 pKa = 11.62YY562 pKa = 10.68FGQLEE567 pKa = 4.44QYY569 pKa = 10.42SITAVAPAGTVKK581 pKa = 10.25VRR583 pKa = 11.84VQSAITCNYY592 pKa = 7.32MKK594 pKa = 10.7LDD596 pKa = 4.12ALCLTATAPLALGDD610 pKa = 4.32RR611 pKa = 11.84VFFDD615 pKa = 4.9LNDD618 pKa = 3.21NGTRR622 pKa = 11.84DD623 pKa = 3.55GSEE626 pKa = 3.68TGLAGVTVKK635 pKa = 10.48LYY637 pKa = 11.12ADD639 pKa = 3.83NNNDD643 pKa = 3.62NIADD647 pKa = 4.08GAALQTTTTDD657 pKa = 2.77ANGFYY662 pKa = 10.5RR663 pKa = 11.84FSNLTAGNYY672 pKa = 8.73IVGVVVPAGYY682 pKa = 10.07RR683 pKa = 11.84SSTLTGVDD691 pKa = 3.23PDD693 pKa = 4.25NNVDD697 pKa = 5.66LDD699 pKa = 4.37DD700 pKa = 4.81NGNNTLISGEE710 pKa = 3.88IRR712 pKa = 11.84GNAVTLSAGSEE723 pKa = 4.01PQEE726 pKa = 3.91SGNFNNTYY734 pKa = 11.03DD735 pKa = 3.87FGFTGTGSIGDD746 pKa = 3.96FVWNDD751 pKa = 2.98LNANGIQDD759 pKa = 3.58AGEE762 pKa = 4.59PGLSGVTVTLTYY774 pKa = 10.48PGGTTVTATTDD785 pKa = 3.12ANGAYY790 pKa = 9.54QFNNLPPGTYY800 pKa = 9.71SISFSTPSGYY810 pKa = 9.81IAGSANQGADD820 pKa = 3.58DD821 pKa = 5.22AKK823 pKa = 11.26DD824 pKa = 3.49SDD826 pKa = 4.47PVSGTVSGIVLTAGQNNTTIDD847 pKa = 3.26AGFVNNQLVLGNFVWYY863 pKa = 9.94DD864 pKa = 3.22KK865 pKa = 11.56DD866 pKa = 3.59NDD868 pKa = 3.97GVQDD872 pKa = 3.57AGEE875 pKa = 4.13PGIAGVTVNLYY886 pKa = 10.68KK887 pKa = 10.72DD888 pKa = 3.69ANGDD892 pKa = 3.93NIADD896 pKa = 4.06GAAVATTTTNASGLYY911 pKa = 10.22SFTGLTAGNYY921 pKa = 8.63IVGVVLPSGYY931 pKa = 10.16AASAVSSATPNNDD944 pKa = 2.47VNTDD948 pKa = 3.41NNGVTTASGEE958 pKa = 4.09VRR960 pKa = 11.84SNYY963 pKa = 8.16ITLSTAGEE971 pKa = 4.23PTTDD975 pKa = 3.79GDD977 pKa = 4.29DD978 pKa = 3.74ANGNLTLDD986 pKa = 3.78FGLRR990 pKa = 11.84GTASIGDD997 pKa = 4.17YY998 pKa = 10.36VWNDD1002 pKa = 3.21LNGNGIQNTGEE1013 pKa = 4.26PGLSGVTVTLTYY1025 pKa = 10.5PDD1027 pKa = 3.87GTTVTTTTDD1036 pKa = 2.77GSGAYY1041 pKa = 10.08LFANLAPGTYY1051 pKa = 9.48SVAFATPNGSIASPSNAGADD1071 pKa = 3.79DD1072 pKa = 4.22TKK1074 pKa = 11.47DD1075 pKa = 3.34SDD1077 pKa = 4.05PVSGVVSGITLTAGQTNTTIDD1098 pKa = 4.02AGFQSNGLNLGNFVWYY1114 pKa = 9.76DD1115 pKa = 3.42RR1116 pKa = 11.84NNNGVQDD1123 pKa = 3.74AGEE1126 pKa = 4.19PGIAGATVKK1135 pKa = 10.58LYY1137 pKa = 11.18ADD1139 pKa = 4.12ANLDD1143 pKa = 4.04NIADD1147 pKa = 4.14GAALQTTTTDD1157 pKa = 2.98ANGIYY1162 pKa = 9.92SFTGLTPGNYY1172 pKa = 8.72IAGVVLPAGYY1182 pKa = 9.66AASSVSSSTPNNDD1195 pKa = 2.62VNTDD1199 pKa = 3.45NNGVTTTSGEE1209 pKa = 3.97VRR1211 pKa = 11.84SNYY1214 pKa = 8.04ITLITASEE1222 pKa = 4.28PTTDD1226 pKa = 3.77GDD1228 pKa = 4.01GSNGNLTLDD1237 pKa = 4.29FGLKK1241 pKa = 8.67GTGSIGDD1248 pKa = 3.92FVWNDD1253 pKa = 3.34LNGDD1257 pKa = 4.58GIQNAGEE1264 pKa = 4.45PGLSAVTVTLTYY1276 pKa = 10.34PGGATATTTTDD1287 pKa = 2.93ANGAYY1292 pKa = 9.87QFTNLAPGTYY1302 pKa = 8.98SVSFTTPSGYY1312 pKa = 9.66IAAPANQGADD1322 pKa = 3.71DD1323 pKa = 5.19AKK1325 pKa = 11.26DD1326 pKa = 3.49SDD1328 pKa = 4.41PVSGTVAGIVLTAGQTNTTIDD1349 pKa = 3.17AGFVSNQLTLGNFVWYY1365 pKa = 9.89DD1366 pKa = 3.22KK1367 pKa = 11.56DD1368 pKa = 3.59NDD1370 pKa = 3.97GVQDD1374 pKa = 3.68AGEE1377 pKa = 4.19LGIAAVTVNLYY1388 pKa = 10.7KK1389 pKa = 10.68DD1390 pKa = 3.68ANGDD1394 pKa = 3.6NVADD1398 pKa = 4.4GAAVATTTTNASGIYY1413 pKa = 9.3NFSGLTPGNYY1423 pKa = 8.78IAGVVLPAGYY1433 pKa = 10.31APSSVSSTTPNNDD1446 pKa = 2.35INTDD1450 pKa = 3.51NNGVTTAAGEE1460 pKa = 4.18VRR1462 pKa = 11.84SNYY1465 pKa = 8.26ITLTTAGEE1473 pKa = 4.16PTTDD1477 pKa = 3.29GDD1479 pKa = 4.07GSNGNLTLDD1488 pKa = 3.73FGLRR1492 pKa = 11.84GTASIGDD1499 pKa = 4.01FVWNDD1504 pKa = 3.34LNGDD1508 pKa = 4.65GIQTAGEE1515 pKa = 3.91PGIAGVTVTLTTAGGTVLEE1534 pKa = 4.64TTTTNASGFYY1544 pKa = 9.96QFSNLLPGTYY1554 pKa = 9.71NVVFATPTGFVASAANAGADD1574 pKa = 3.82DD1575 pKa = 4.62TKK1577 pKa = 11.47DD1578 pKa = 3.34SDD1580 pKa = 4.25PVSGTVTGVVLNADD1594 pKa = 3.93EE1595 pKa = 4.79NDD1597 pKa = 3.53NTVDD1601 pKa = 5.54AGFQSNQLKK1610 pKa = 9.54VGNYY1614 pKa = 6.99VWYY1617 pKa = 9.93DD1618 pKa = 3.23KK1619 pKa = 11.42NNNGFQDD1626 pKa = 3.51ADD1628 pKa = 3.53EE1629 pKa = 4.4TGIAGVSVKK1638 pKa = 10.74LYY1640 pKa = 11.09VDD1642 pKa = 4.11ADD1644 pKa = 3.84ANNIPDD1650 pKa = 3.77AAAIASTTTDD1660 pKa = 2.99ANGLYY1665 pKa = 9.9SFSGLTPGNYY1675 pKa = 8.92IVGITAPSGYY1685 pKa = 11.24AMAFTTLSSLLPNNDD1700 pKa = 3.19NNTDD1704 pKa = 3.56NNGITIIGTEE1714 pKa = 3.68ITSNYY1719 pKa = 7.36ITLITNSEE1727 pKa = 4.26PVTDD1731 pKa = 3.88GDD1733 pKa = 4.39GNNGNLTLDD1742 pKa = 4.21FGLKK1746 pKa = 8.67GTGSIGDD1753 pKa = 4.43LVWDD1757 pKa = 4.67DD1758 pKa = 4.79LNANGKK1764 pKa = 9.13QDD1766 pKa = 3.36AGEE1769 pKa = 4.09PGIAGVTVTLTYY1781 pKa = 10.33PGGSTTSVITDD1792 pKa = 3.18ATGTYY1797 pKa = 10.08LFSNLAPGSYY1807 pKa = 9.83SVSFDD1812 pKa = 3.26KK1813 pKa = 11.0PLGYY1817 pKa = 9.69IEE1819 pKa = 4.08TTANQVADD1827 pKa = 3.93DD1828 pKa = 4.4TKK1830 pKa = 11.37DD1831 pKa = 3.23SDD1833 pKa = 4.41VISGSVSNVTLTAGQTNFTVDD1854 pKa = 2.67AGYY1857 pKa = 10.76YY1858 pKa = 9.26KK1859 pKa = 10.22PASIGDD1865 pKa = 3.92FVWNDD1870 pKa = 2.98LNANGIQDD1878 pKa = 3.55AGEE1881 pKa = 4.11PGIAGATVNLTGTDD1895 pKa = 3.27ATGAAVNLTVSTEE1908 pKa = 3.88ADD1910 pKa = 3.16GSYY1913 pKa = 10.65SFSSLLPGTYY1923 pKa = 9.32NVSFVTPAGYY1933 pKa = 7.82VTATSNAGADD1943 pKa = 3.81DD1944 pKa = 4.36TKK1946 pKa = 11.47DD1947 pKa = 3.33SDD1949 pKa = 4.24PVSGAVNGITLISGQNNATVDD1970 pKa = 3.24AGFIQQTPIVLSGHH1984 pKa = 5.79VFYY1987 pKa = 10.79DD1988 pKa = 3.77DD1989 pKa = 4.81NGMSDD1994 pKa = 3.55GFVNGTSVIPSGINAVLYY2012 pKa = 10.59DD2013 pKa = 4.2PLTNTVVAVQPVPGYY2028 pKa = 8.42GQPGAGTFSFNVLTYY2043 pKa = 8.32STYY2046 pKa = 10.69RR2047 pKa = 11.84VLITTANPAVGSQAPVTSVLPASFRR2072 pKa = 11.84STGEE2076 pKa = 4.14TIGTAAGNDD2085 pKa = 3.62GTIDD2089 pKa = 3.72GKK2091 pKa = 11.12SSIINTTNQNIIEE2104 pKa = 4.26VNFAIKK2110 pKa = 10.02RR2111 pKa = 11.84VVLIVVDD2118 pKa = 4.03
MM1 pKa = 7.29KK2 pKa = 10.46QFLPYY7 pKa = 9.97KK8 pKa = 8.45RR9 pKa = 11.84NCMGAAIKK17 pKa = 10.24NLHH20 pKa = 6.56LLFTLSIALFVSTNVANAQCTCTGNIVVNPSFEE53 pKa = 4.39SGTTGWSWSGGSFNAGTGAIACGSKK78 pKa = 10.49SGDD81 pKa = 3.42FQISNTSSNWVSQTIGTDD99 pKa = 3.72LAPGTKK105 pKa = 9.47INASVYY111 pKa = 10.77AGTHH115 pKa = 5.07NNSYY119 pKa = 8.75YY120 pKa = 10.35HH121 pKa = 5.9QVAIDD126 pKa = 3.92FFDD129 pKa = 5.82ANWNWISNSKK139 pKa = 10.38VEE141 pKa = 4.21VNKK144 pKa = 10.1VLSTSPVGPQLYY156 pKa = 8.27TWSGTVPVGTKK167 pKa = 7.39YY168 pKa = 9.61TNVSFSGNGDD178 pKa = 3.84WIKK181 pKa = 10.2TDD183 pKa = 3.21QWCVTLTPPAGGSIGDD199 pKa = 4.23RR200 pKa = 11.84VWLDD204 pKa = 3.06ADD206 pKa = 4.07GDD208 pKa = 4.27GLQDD212 pKa = 3.4ASEE215 pKa = 4.26TSGIAGVTVQLKK227 pKa = 10.25NSLGSVIATTTTNASGNYY245 pKa = 9.76LFTGLAAGNYY255 pKa = 6.93TVVFPTSISGAVVTPQNVGLDD276 pKa = 3.97DD277 pKa = 4.96NVDD280 pKa = 3.47SDD282 pKa = 4.49ASQTTGATSTITLATNQNITNVDD305 pKa = 3.06AGYY308 pKa = 10.83CPTTLSLGNRR318 pKa = 11.84VWYY321 pKa = 7.73DD322 pKa = 3.48TNNDD326 pKa = 4.14GINSNEE332 pKa = 3.8NGIPNLTVYY341 pKa = 10.55LYY343 pKa = 11.09KK344 pKa = 10.48DD345 pKa = 3.74DD346 pKa = 5.55NNDD349 pKa = 3.19NVADD353 pKa = 3.93AAAIATAVTDD363 pKa = 3.82ANGYY367 pKa = 10.47YY368 pKa = 9.01MFSNLAPGNYY378 pKa = 8.66IVGAVTPAGYY388 pKa = 9.23MSSLVNGGDD397 pKa = 3.46PDD399 pKa = 5.01DD400 pKa = 5.17NNNSDD405 pKa = 4.55DD406 pKa = 3.97NGEE409 pKa = 4.24VTVGAEE415 pKa = 4.05TRR417 pKa = 11.84GLAITLVAGTEE428 pKa = 3.99PLVDD432 pKa = 3.89GNTNSNSNITYY443 pKa = 10.52DD444 pKa = 4.2FGFLPDD450 pKa = 4.37CNCTSSSSNLLINGSFEE467 pKa = 4.18NGTTGWSASGGSVSSGTGYY486 pKa = 9.57VACGAANGFNSWSSGTSRR504 pKa = 11.84VWQDD508 pKa = 2.87VAVAAGSTVVFKK520 pKa = 11.02GFAGVHH526 pKa = 4.68TAGLTCSPKK535 pKa = 10.61LSLIFYY541 pKa = 9.14NSSNTVISQNDD552 pKa = 3.44VVVTRR557 pKa = 11.84DD558 pKa = 2.69VDD560 pKa = 3.52KK561 pKa = 11.62YY562 pKa = 10.68FGQLEE567 pKa = 4.44QYY569 pKa = 10.42SITAVAPAGTVKK581 pKa = 10.25VRR583 pKa = 11.84VQSAITCNYY592 pKa = 7.32MKK594 pKa = 10.7LDD596 pKa = 4.12ALCLTATAPLALGDD610 pKa = 4.32RR611 pKa = 11.84VFFDD615 pKa = 4.9LNDD618 pKa = 3.21NGTRR622 pKa = 11.84DD623 pKa = 3.55GSEE626 pKa = 3.68TGLAGVTVKK635 pKa = 10.48LYY637 pKa = 11.12ADD639 pKa = 3.83NNNDD643 pKa = 3.62NIADD647 pKa = 4.08GAALQTTTTDD657 pKa = 2.77ANGFYY662 pKa = 10.5RR663 pKa = 11.84FSNLTAGNYY672 pKa = 8.73IVGVVVPAGYY682 pKa = 10.07RR683 pKa = 11.84SSTLTGVDD691 pKa = 3.23PDD693 pKa = 4.25NNVDD697 pKa = 5.66LDD699 pKa = 4.37DD700 pKa = 4.81NGNNTLISGEE710 pKa = 3.88IRR712 pKa = 11.84GNAVTLSAGSEE723 pKa = 4.01PQEE726 pKa = 3.91SGNFNNTYY734 pKa = 11.03DD735 pKa = 3.87FGFTGTGSIGDD746 pKa = 3.96FVWNDD751 pKa = 2.98LNANGIQDD759 pKa = 3.58AGEE762 pKa = 4.59PGLSGVTVTLTYY774 pKa = 10.48PGGTTVTATTDD785 pKa = 3.12ANGAYY790 pKa = 9.54QFNNLPPGTYY800 pKa = 9.71SISFSTPSGYY810 pKa = 9.81IAGSANQGADD820 pKa = 3.58DD821 pKa = 5.22AKK823 pKa = 11.26DD824 pKa = 3.49SDD826 pKa = 4.47PVSGTVSGIVLTAGQNNTTIDD847 pKa = 3.26AGFVNNQLVLGNFVWYY863 pKa = 9.94DD864 pKa = 3.22KK865 pKa = 11.56DD866 pKa = 3.59NDD868 pKa = 3.97GVQDD872 pKa = 3.57AGEE875 pKa = 4.13PGIAGVTVNLYY886 pKa = 10.68KK887 pKa = 10.72DD888 pKa = 3.69ANGDD892 pKa = 3.93NIADD896 pKa = 4.06GAAVATTTTNASGLYY911 pKa = 10.22SFTGLTAGNYY921 pKa = 8.63IVGVVLPSGYY931 pKa = 10.16AASAVSSATPNNDD944 pKa = 2.47VNTDD948 pKa = 3.41NNGVTTASGEE958 pKa = 4.09VRR960 pKa = 11.84SNYY963 pKa = 8.16ITLSTAGEE971 pKa = 4.23PTTDD975 pKa = 3.79GDD977 pKa = 4.29DD978 pKa = 3.74ANGNLTLDD986 pKa = 3.78FGLRR990 pKa = 11.84GTASIGDD997 pKa = 4.17YY998 pKa = 10.36VWNDD1002 pKa = 3.21LNGNGIQNTGEE1013 pKa = 4.26PGLSGVTVTLTYY1025 pKa = 10.5PDD1027 pKa = 3.87GTTVTTTTDD1036 pKa = 2.77GSGAYY1041 pKa = 10.08LFANLAPGTYY1051 pKa = 9.48SVAFATPNGSIASPSNAGADD1071 pKa = 3.79DD1072 pKa = 4.22TKK1074 pKa = 11.47DD1075 pKa = 3.34SDD1077 pKa = 4.05PVSGVVSGITLTAGQTNTTIDD1098 pKa = 4.02AGFQSNGLNLGNFVWYY1114 pKa = 9.76DD1115 pKa = 3.42RR1116 pKa = 11.84NNNGVQDD1123 pKa = 3.74AGEE1126 pKa = 4.19PGIAGATVKK1135 pKa = 10.58LYY1137 pKa = 11.18ADD1139 pKa = 4.12ANLDD1143 pKa = 4.04NIADD1147 pKa = 4.14GAALQTTTTDD1157 pKa = 2.98ANGIYY1162 pKa = 9.92SFTGLTPGNYY1172 pKa = 8.72IAGVVLPAGYY1182 pKa = 9.66AASSVSSSTPNNDD1195 pKa = 2.62VNTDD1199 pKa = 3.45NNGVTTTSGEE1209 pKa = 3.97VRR1211 pKa = 11.84SNYY1214 pKa = 8.04ITLITASEE1222 pKa = 4.28PTTDD1226 pKa = 3.77GDD1228 pKa = 4.01GSNGNLTLDD1237 pKa = 4.29FGLKK1241 pKa = 8.67GTGSIGDD1248 pKa = 3.92FVWNDD1253 pKa = 3.34LNGDD1257 pKa = 4.58GIQNAGEE1264 pKa = 4.45PGLSAVTVTLTYY1276 pKa = 10.34PGGATATTTTDD1287 pKa = 2.93ANGAYY1292 pKa = 9.87QFTNLAPGTYY1302 pKa = 8.98SVSFTTPSGYY1312 pKa = 9.66IAAPANQGADD1322 pKa = 3.71DD1323 pKa = 5.19AKK1325 pKa = 11.26DD1326 pKa = 3.49SDD1328 pKa = 4.41PVSGTVAGIVLTAGQTNTTIDD1349 pKa = 3.17AGFVSNQLTLGNFVWYY1365 pKa = 9.89DD1366 pKa = 3.22KK1367 pKa = 11.56DD1368 pKa = 3.59NDD1370 pKa = 3.97GVQDD1374 pKa = 3.68AGEE1377 pKa = 4.19LGIAAVTVNLYY1388 pKa = 10.7KK1389 pKa = 10.68DD1390 pKa = 3.68ANGDD1394 pKa = 3.6NVADD1398 pKa = 4.4GAAVATTTTNASGIYY1413 pKa = 9.3NFSGLTPGNYY1423 pKa = 8.78IAGVVLPAGYY1433 pKa = 10.31APSSVSSTTPNNDD1446 pKa = 2.35INTDD1450 pKa = 3.51NNGVTTAAGEE1460 pKa = 4.18VRR1462 pKa = 11.84SNYY1465 pKa = 8.26ITLTTAGEE1473 pKa = 4.16PTTDD1477 pKa = 3.29GDD1479 pKa = 4.07GSNGNLTLDD1488 pKa = 3.73FGLRR1492 pKa = 11.84GTASIGDD1499 pKa = 4.01FVWNDD1504 pKa = 3.34LNGDD1508 pKa = 4.65GIQTAGEE1515 pKa = 3.91PGIAGVTVTLTTAGGTVLEE1534 pKa = 4.64TTTTNASGFYY1544 pKa = 9.96QFSNLLPGTYY1554 pKa = 9.71NVVFATPTGFVASAANAGADD1574 pKa = 3.82DD1575 pKa = 4.62TKK1577 pKa = 11.47DD1578 pKa = 3.34SDD1580 pKa = 4.25PVSGTVTGVVLNADD1594 pKa = 3.93EE1595 pKa = 4.79NDD1597 pKa = 3.53NTVDD1601 pKa = 5.54AGFQSNQLKK1610 pKa = 9.54VGNYY1614 pKa = 6.99VWYY1617 pKa = 9.93DD1618 pKa = 3.23KK1619 pKa = 11.42NNNGFQDD1626 pKa = 3.51ADD1628 pKa = 3.53EE1629 pKa = 4.4TGIAGVSVKK1638 pKa = 10.74LYY1640 pKa = 11.09VDD1642 pKa = 4.11ADD1644 pKa = 3.84ANNIPDD1650 pKa = 3.77AAAIASTTTDD1660 pKa = 2.99ANGLYY1665 pKa = 9.9SFSGLTPGNYY1675 pKa = 8.92IVGITAPSGYY1685 pKa = 11.24AMAFTTLSSLLPNNDD1700 pKa = 3.19NNTDD1704 pKa = 3.56NNGITIIGTEE1714 pKa = 3.68ITSNYY1719 pKa = 7.36ITLITNSEE1727 pKa = 4.26PVTDD1731 pKa = 3.88GDD1733 pKa = 4.39GNNGNLTLDD1742 pKa = 4.21FGLKK1746 pKa = 8.67GTGSIGDD1753 pKa = 4.43LVWDD1757 pKa = 4.67DD1758 pKa = 4.79LNANGKK1764 pKa = 9.13QDD1766 pKa = 3.36AGEE1769 pKa = 4.09PGIAGVTVTLTYY1781 pKa = 10.33PGGSTTSVITDD1792 pKa = 3.18ATGTYY1797 pKa = 10.08LFSNLAPGSYY1807 pKa = 9.83SVSFDD1812 pKa = 3.26KK1813 pKa = 11.0PLGYY1817 pKa = 9.69IEE1819 pKa = 4.08TTANQVADD1827 pKa = 3.93DD1828 pKa = 4.4TKK1830 pKa = 11.37DD1831 pKa = 3.23SDD1833 pKa = 4.41VISGSVSNVTLTAGQTNFTVDD1854 pKa = 2.67AGYY1857 pKa = 10.76YY1858 pKa = 9.26KK1859 pKa = 10.22PASIGDD1865 pKa = 3.92FVWNDD1870 pKa = 2.98LNANGIQDD1878 pKa = 3.55AGEE1881 pKa = 4.11PGIAGATVNLTGTDD1895 pKa = 3.27ATGAAVNLTVSTEE1908 pKa = 3.88ADD1910 pKa = 3.16GSYY1913 pKa = 10.65SFSSLLPGTYY1923 pKa = 9.32NVSFVTPAGYY1933 pKa = 7.82VTATSNAGADD1943 pKa = 3.81DD1944 pKa = 4.36TKK1946 pKa = 11.47DD1947 pKa = 3.33SDD1949 pKa = 4.24PVSGAVNGITLISGQNNATVDD1970 pKa = 3.24AGFIQQTPIVLSGHH1984 pKa = 5.79VFYY1987 pKa = 10.79DD1988 pKa = 3.77DD1989 pKa = 4.81NGMSDD1994 pKa = 3.55GFVNGTSVIPSGINAVLYY2012 pKa = 10.59DD2013 pKa = 4.2PLTNTVVAVQPVPGYY2028 pKa = 8.42GQPGAGTFSFNVLTYY2043 pKa = 8.32STYY2046 pKa = 10.69RR2047 pKa = 11.84VLITTANPAVGSQAPVTSVLPASFRR2072 pKa = 11.84STGEE2076 pKa = 4.14TIGTAAGNDD2085 pKa = 3.62GTIDD2089 pKa = 3.72GKK2091 pKa = 11.12SSIINTTNQNIIEE2104 pKa = 4.26VNFAIKK2110 pKa = 10.02RR2111 pKa = 11.84VVLIVVDD2118 pKa = 4.03
Molecular weight: 216.14 kDa
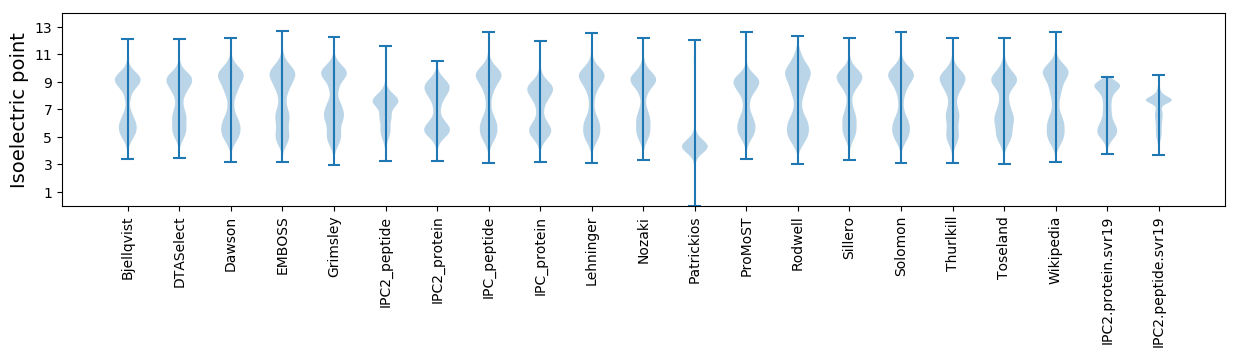
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V1M7T8|A0A4V1M7T8_9BACT Alpha-E domain-containing protein OS=Lacibacter luteus OX=2508719 GN=ESA94_00335 PE=4 SV=1
MM1 pKa = 7.99RR2 pKa = 11.84SYY4 pKa = 10.71FKK6 pKa = 10.46QWTGMRR12 pKa = 11.84LLRR15 pKa = 11.84LLMGVVAIVQGIASPTPLLWVIGAVLLVQAFLNIGCMDD53 pKa = 4.36SSCNIPVQQQKK64 pKa = 7.25MHH66 pKa = 6.54RR67 pKa = 3.77
MM1 pKa = 7.99RR2 pKa = 11.84SYY4 pKa = 10.71FKK6 pKa = 10.46QWTGMRR12 pKa = 11.84LLRR15 pKa = 11.84LLMGVVAIVQGIASPTPLLWVIGAVLLVQAFLNIGCMDD53 pKa = 4.36SSCNIPVQQQKK64 pKa = 7.25MHH66 pKa = 6.54RR67 pKa = 3.77
Molecular weight: 7.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1521579 |
15 |
3128 |
364.0 |
40.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.746 ± 0.034 | 0.854 ± 0.013 |
5.036 ± 0.026 | 5.643 ± 0.038 |
5.035 ± 0.027 | 6.787 ± 0.037 |
1.829 ± 0.017 | 6.668 ± 0.029 |
7.036 ± 0.039 | 9.131 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.323 ± 0.021 | 5.76 ± 0.04 |
3.726 ± 0.021 | 3.963 ± 0.023 |
3.863 ± 0.023 | 6.241 ± 0.032 |
6.185 ± 0.048 | 6.697 ± 0.025 |
1.375 ± 0.014 | 4.102 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
