
Pigeon adenovirus 1
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Tectiliviricetes; Rowavirales; Adenoviridae; Aviadenovirus; Pigeon aviadenovirus A
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 37 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
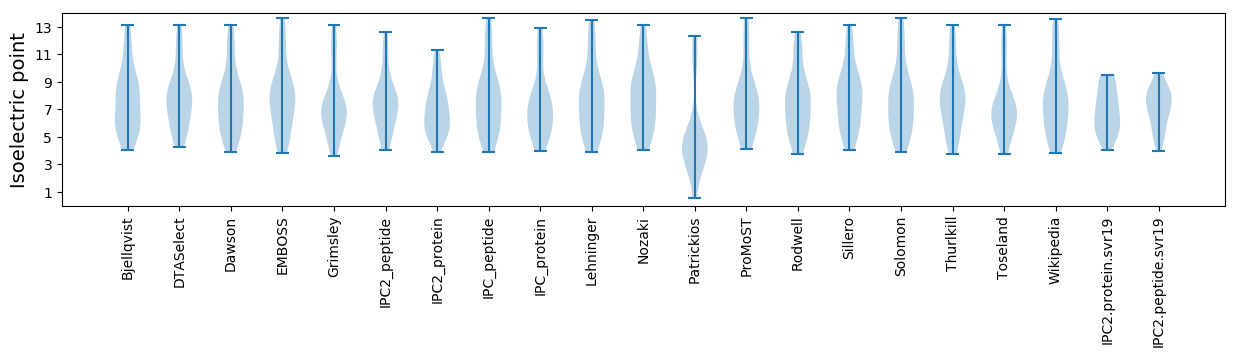
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|X5M219|X5M219_9ADEN Protein ORF62 OS=Pigeon adenovirus 1 OX=764030 GN=ORF62 PE=4 SV=1
MM1 pKa = 7.37PQPSNVAVVAPKK13 pKa = 9.49TKK15 pKa = 10.35PEE17 pKa = 3.97APRR20 pKa = 11.84KK21 pKa = 8.71RR22 pKa = 11.84QLSAAALAAEE32 pKa = 4.25QQQAEE37 pKa = 4.02RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84KK42 pKa = 9.63RR43 pKa = 11.84LRR45 pKa = 11.84RR46 pKa = 11.84DD47 pKa = 3.03ATDD50 pKa = 3.94PDD52 pKa = 4.26LNLVYY57 pKa = 9.81PFWYY61 pKa = 8.98TVEE64 pKa = 4.68DD65 pKa = 3.62PHH67 pKa = 8.45VVNPPFLAPTGPLYY81 pKa = 10.78DD82 pKa = 4.93DD83 pKa = 4.55NGNLNIRR90 pKa = 11.84LTPPVTLVEE99 pKa = 4.82DD100 pKa = 4.59GVGLAYY106 pKa = 10.29DD107 pKa = 3.55QSLKK111 pKa = 10.98VSDD114 pKa = 3.86ATGAAAGVLGVQVDD128 pKa = 3.99TEE130 pKa = 4.56GPLTVTEE137 pKa = 5.51DD138 pKa = 3.84GLDD141 pKa = 3.78LNTDD145 pKa = 3.34DD146 pKa = 5.5SLAVNDD152 pKa = 4.05DD153 pKa = 3.12WQLGVRR159 pKa = 11.84FAEE162 pKa = 4.0NQPFSVTTAGVSLLVDD178 pKa = 3.66DD179 pKa = 4.98TLLIAAADD187 pKa = 3.98EE188 pKa = 4.3EE189 pKa = 5.24TPTSYY194 pKa = 11.23EE195 pKa = 3.98LGVHH199 pKa = 6.84LNQNGPITADD209 pKa = 3.1ADD211 pKa = 4.66GIDD214 pKa = 4.89LEE216 pKa = 4.79VDD218 pKa = 3.21NQTLQVTTNEE228 pKa = 3.85QSQGVLAVKK237 pKa = 10.29LKK239 pKa = 10.02QQGGLSAGTSGIGINYY255 pKa = 9.32DD256 pKa = 3.29NEE258 pKa = 4.16GALVVSDD265 pKa = 3.86NTLQLKK271 pKa = 10.29IGTDD275 pKa = 3.66QPFSTTNGLSLNYY288 pKa = 9.78DD289 pKa = 3.6PNSFTVSGVTTSGTTTTMSQLAVKK313 pKa = 9.75VASSNPLGKK322 pKa = 10.31DD323 pKa = 3.03NSGLKK328 pKa = 10.0LVYY331 pKa = 9.06NTQDD335 pKa = 3.17FFDD338 pKa = 5.5DD339 pKa = 3.81NSSGLSAVTPIQYY352 pKa = 10.5LSPYY356 pKa = 8.05CTYY359 pKa = 10.95SAGSPGLDD367 pKa = 2.94TFDD370 pKa = 3.27VTVRR374 pKa = 11.84SSSGEE379 pKa = 3.66NWTIAAYY386 pKa = 10.14VVIANSGGICNGILYY401 pKa = 9.8LHH403 pKa = 7.09FDD405 pKa = 3.72KK406 pKa = 11.14KK407 pKa = 10.92QLGTLGTGQGGEE419 pKa = 3.95TGNYY423 pKa = 7.95LRR425 pKa = 11.84CAIVVNPSGNPGGNHH440 pKa = 6.48SNFSNPRR447 pKa = 11.84WWPEE451 pKa = 3.37GSNSFLTPPGPPNAPPVNAVVSAIGRR477 pKa = 11.84NNWYY481 pKa = 10.15YY482 pKa = 9.84GPPWRR487 pKa = 11.84GVEE490 pKa = 4.11VKK492 pKa = 10.59LKK494 pKa = 10.62SRR496 pKa = 11.84STNTIGSTTTVTNTDD511 pKa = 2.66AVAYY515 pKa = 6.92YY516 pKa = 10.58LPATAQGSNGNPNVIYY532 pKa = 9.68FIYY535 pKa = 9.99EE536 pKa = 4.13VKK538 pKa = 10.44FDD540 pKa = 3.9WYY542 pKa = 11.35NGTTNNGMGVQTSDD556 pKa = 5.04PIPFSYY562 pKa = 10.36LGSLPQFPRR571 pKa = 11.84NSTGTISTTTTTTTTVITPALLPDD595 pKa = 3.99VVPAPGPEE603 pKa = 4.5PGTLPAPEE611 pKa = 5.38AGEE614 pKa = 4.28TQDD617 pKa = 3.41TAEE620 pKa = 5.17DD621 pKa = 4.51LPHH624 pKa = 6.48QPPEE628 pKa = 4.32DD629 pKa = 3.49PALEE633 pKa = 4.19VEE635 pKa = 4.35PDD637 pKa = 3.39EE638 pKa = 5.87DD639 pKa = 3.65PALEE643 pKa = 4.22VEE645 pKa = 4.46APEE648 pKa = 4.52DD649 pKa = 3.21AAAEE653 pKa = 4.3VPVVDD658 pKa = 5.07DD659 pKa = 3.99AALEE663 pKa = 4.11EE664 pKa = 4.72VPLEE668 pKa = 4.59DD669 pKa = 4.54PSEE672 pKa = 4.28GSPAEE677 pKa = 4.29DD678 pKa = 2.82AALPEE683 pKa = 4.29EE684 pKa = 4.83DD685 pKa = 4.47VAPEE689 pKa = 3.9DD690 pKa = 3.64TSVVDD695 pKa = 4.76EE696 pKa = 5.29LPPPPPQEE704 pKa = 4.28PPAPDD709 pKa = 4.04PEE711 pKa = 4.96PPVTPPAEE719 pKa = 4.17QPSLVIVDD727 pKa = 5.57FVADD731 pKa = 4.28PDD733 pKa = 4.55PYY735 pKa = 11.15LQFPQDD741 pKa = 3.62HH742 pKa = 4.94QHH744 pKa = 6.81DD745 pKa = 4.56RR746 pKa = 11.84IILL749 pKa = 3.88
MM1 pKa = 7.37PQPSNVAVVAPKK13 pKa = 9.49TKK15 pKa = 10.35PEE17 pKa = 3.97APRR20 pKa = 11.84KK21 pKa = 8.71RR22 pKa = 11.84QLSAAALAAEE32 pKa = 4.25QQQAEE37 pKa = 4.02RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84KK42 pKa = 9.63RR43 pKa = 11.84LRR45 pKa = 11.84RR46 pKa = 11.84DD47 pKa = 3.03ATDD50 pKa = 3.94PDD52 pKa = 4.26LNLVYY57 pKa = 9.81PFWYY61 pKa = 8.98TVEE64 pKa = 4.68DD65 pKa = 3.62PHH67 pKa = 8.45VVNPPFLAPTGPLYY81 pKa = 10.78DD82 pKa = 4.93DD83 pKa = 4.55NGNLNIRR90 pKa = 11.84LTPPVTLVEE99 pKa = 4.82DD100 pKa = 4.59GVGLAYY106 pKa = 10.29DD107 pKa = 3.55QSLKK111 pKa = 10.98VSDD114 pKa = 3.86ATGAAAGVLGVQVDD128 pKa = 3.99TEE130 pKa = 4.56GPLTVTEE137 pKa = 5.51DD138 pKa = 3.84GLDD141 pKa = 3.78LNTDD145 pKa = 3.34DD146 pKa = 5.5SLAVNDD152 pKa = 4.05DD153 pKa = 3.12WQLGVRR159 pKa = 11.84FAEE162 pKa = 4.0NQPFSVTTAGVSLLVDD178 pKa = 3.66DD179 pKa = 4.98TLLIAAADD187 pKa = 3.98EE188 pKa = 4.3EE189 pKa = 5.24TPTSYY194 pKa = 11.23EE195 pKa = 3.98LGVHH199 pKa = 6.84LNQNGPITADD209 pKa = 3.1ADD211 pKa = 4.66GIDD214 pKa = 4.89LEE216 pKa = 4.79VDD218 pKa = 3.21NQTLQVTTNEE228 pKa = 3.85QSQGVLAVKK237 pKa = 10.29LKK239 pKa = 10.02QQGGLSAGTSGIGINYY255 pKa = 9.32DD256 pKa = 3.29NEE258 pKa = 4.16GALVVSDD265 pKa = 3.86NTLQLKK271 pKa = 10.29IGTDD275 pKa = 3.66QPFSTTNGLSLNYY288 pKa = 9.78DD289 pKa = 3.6PNSFTVSGVTTSGTTTTMSQLAVKK313 pKa = 9.75VASSNPLGKK322 pKa = 10.31DD323 pKa = 3.03NSGLKK328 pKa = 10.0LVYY331 pKa = 9.06NTQDD335 pKa = 3.17FFDD338 pKa = 5.5DD339 pKa = 3.81NSSGLSAVTPIQYY352 pKa = 10.5LSPYY356 pKa = 8.05CTYY359 pKa = 10.95SAGSPGLDD367 pKa = 2.94TFDD370 pKa = 3.27VTVRR374 pKa = 11.84SSSGEE379 pKa = 3.66NWTIAAYY386 pKa = 10.14VVIANSGGICNGILYY401 pKa = 9.8LHH403 pKa = 7.09FDD405 pKa = 3.72KK406 pKa = 11.14KK407 pKa = 10.92QLGTLGTGQGGEE419 pKa = 3.95TGNYY423 pKa = 7.95LRR425 pKa = 11.84CAIVVNPSGNPGGNHH440 pKa = 6.48SNFSNPRR447 pKa = 11.84WWPEE451 pKa = 3.37GSNSFLTPPGPPNAPPVNAVVSAIGRR477 pKa = 11.84NNWYY481 pKa = 10.15YY482 pKa = 9.84GPPWRR487 pKa = 11.84GVEE490 pKa = 4.11VKK492 pKa = 10.59LKK494 pKa = 10.62SRR496 pKa = 11.84STNTIGSTTTVTNTDD511 pKa = 2.66AVAYY515 pKa = 6.92YY516 pKa = 10.58LPATAQGSNGNPNVIYY532 pKa = 9.68FIYY535 pKa = 9.99EE536 pKa = 4.13VKK538 pKa = 10.44FDD540 pKa = 3.9WYY542 pKa = 11.35NGTTNNGMGVQTSDD556 pKa = 5.04PIPFSYY562 pKa = 10.36LGSLPQFPRR571 pKa = 11.84NSTGTISTTTTTTTTVITPALLPDD595 pKa = 3.99VVPAPGPEE603 pKa = 4.5PGTLPAPEE611 pKa = 5.38AGEE614 pKa = 4.28TQDD617 pKa = 3.41TAEE620 pKa = 5.17DD621 pKa = 4.51LPHH624 pKa = 6.48QPPEE628 pKa = 4.32DD629 pKa = 3.49PALEE633 pKa = 4.19VEE635 pKa = 4.35PDD637 pKa = 3.39EE638 pKa = 5.87DD639 pKa = 3.65PALEE643 pKa = 4.22VEE645 pKa = 4.46APEE648 pKa = 4.52DD649 pKa = 3.21AAAEE653 pKa = 4.3VPVVDD658 pKa = 5.07DD659 pKa = 3.99AALEE663 pKa = 4.11EE664 pKa = 4.72VPLEE668 pKa = 4.59DD669 pKa = 4.54PSEE672 pKa = 4.28GSPAEE677 pKa = 4.29DD678 pKa = 2.82AALPEE683 pKa = 4.29EE684 pKa = 4.83DD685 pKa = 4.47VAPEE689 pKa = 3.9DD690 pKa = 3.64TSVVDD695 pKa = 4.76EE696 pKa = 5.29LPPPPPQEE704 pKa = 4.28PPAPDD709 pKa = 4.04PEE711 pKa = 4.96PPVTPPAEE719 pKa = 4.17QPSLVIVDD727 pKa = 5.57FVADD731 pKa = 4.28PDD733 pKa = 4.55PYY735 pKa = 11.15LQFPQDD741 pKa = 3.62HH742 pKa = 4.94QHH744 pKa = 6.81DD745 pKa = 4.56RR746 pKa = 11.84IILL749 pKa = 3.88
Molecular weight: 79.4 kDa
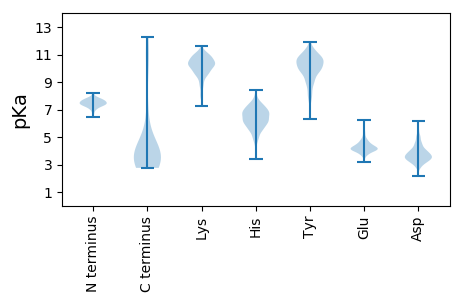
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|X5LU16|X5LU16_9ADEN Hexon assembly protein 100K OS=Pigeon adenovirus 1 OX=764030 GN=100K PE=4 SV=1
MM1 pKa = 7.74SILISPSDD9 pKa = 3.46NRR11 pKa = 11.84GWGMRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84PRR20 pKa = 11.84SSMRR24 pKa = 11.84GVGSTGRR31 pKa = 11.84LVLRR35 pKa = 11.84RR36 pKa = 11.84LLGLGTTTSRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84TRR53 pKa = 11.84RR54 pKa = 11.84VRR56 pKa = 11.84GGAARR61 pKa = 11.84RR62 pKa = 11.84STTSTTRR69 pKa = 11.84VIAVRR74 pKa = 11.84TTRR77 pKa = 11.84RR78 pKa = 11.84RR79 pKa = 11.84RR80 pKa = 11.84RR81 pKa = 11.84RR82 pKa = 3.29
MM1 pKa = 7.74SILISPSDD9 pKa = 3.46NRR11 pKa = 11.84GWGMRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84PRR20 pKa = 11.84SSMRR24 pKa = 11.84GVGSTGRR31 pKa = 11.84LVLRR35 pKa = 11.84RR36 pKa = 11.84LLGLGTTTSRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84TRR53 pKa = 11.84RR54 pKa = 11.84VRR56 pKa = 11.84GGAARR61 pKa = 11.84RR62 pKa = 11.84STTSTTRR69 pKa = 11.84VIAVRR74 pKa = 11.84TTRR77 pKa = 11.84RR78 pKa = 11.84RR79 pKa = 11.84RR80 pKa = 11.84RR81 pKa = 11.84RR82 pKa = 3.29
Molecular weight: 9.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13690 |
37 |
1571 |
370.0 |
41.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.072 ± 0.509 | 1.819 ± 0.248 |
5.398 ± 0.282 | 5.778 ± 0.393 |
3.937 ± 0.253 | 6.275 ± 0.339 |
2.462 ± 0.253 | 2.476 ± 0.168 |
2.345 ± 0.185 | 9.905 ± 0.432 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.833 ± 0.152 | 3.492 ± 0.492 |
7.714 ± 0.458 | 3.901 ± 0.262 |
8.787 ± 0.666 | 6.45 ± 0.297 |
6.326 ± 0.664 | 6.896 ± 0.286 |
1.461 ± 0.153 | 3.674 ± 0.292 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
