
Fusarium vanettenii (strain ATCC MYA-4622 / CBS 123669 / FGSC 9596 / NRRL 45880 / 77-13-4) (Fusarium solani subsp. pisi)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Hypocreales; Nectriaceae; Fusarium; Fusarium solani species complex; Fusarium vanettenii
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
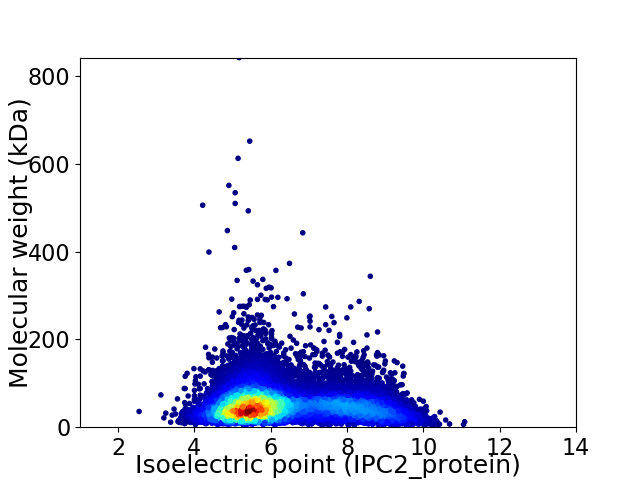
Virtual 2D-PAGE plot for 15709 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C7YQ68|C7YQ68_FUSV7 Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit OST2 OS=Fusarium vanettenii (strain ATCC MYA-4622 / CBS 123669 / FGSC 9596 / NRRL 45880 / 77-13-4) OX=660122 GN=NECHADRAFT_78998 PE=3 SV=1
MM1 pKa = 7.64RR2 pKa = 11.84VLLSRR7 pKa = 11.84LIALAIFLPLGVGFTIVTNSDD28 pKa = 3.13GNTLGNAMMPGSGITLVSGTYY49 pKa = 9.28TGASVAAGTFSDD61 pKa = 4.6GPFGISSGTILTSGRR76 pKa = 11.84ADD78 pKa = 3.4GATVAGSDD86 pKa = 4.12NDD88 pKa = 3.78NGQPGYY94 pKa = 10.14MGGNTFDD101 pKa = 4.03AAVLSLDD108 pKa = 3.41ITIAPPFTGFEE119 pKa = 4.32LEE121 pKa = 4.66FAFASNDD128 pKa = 3.38YY129 pKa = 10.93GGQQQFDD136 pKa = 4.38DD137 pKa = 4.17EE138 pKa = 4.83AADD141 pKa = 3.58NTRR144 pKa = 11.84TIFDD148 pKa = 4.14LVNTFMDD155 pKa = 4.3PPYY158 pKa = 10.57SINPPNSLTAYY169 pKa = 9.26DD170 pKa = 4.01KK171 pKa = 11.2SSPPVLIGFATGSGAFNVKK190 pKa = 9.76IAVYY194 pKa = 10.01DD195 pKa = 4.06LNDD198 pKa = 3.64GFEE201 pKa = 4.71DD202 pKa = 3.51SAALIRR208 pKa = 11.84MRR210 pKa = 11.84GCEE213 pKa = 3.82NCAAGPVINYY223 pKa = 5.8EE224 pKa = 4.35TKK226 pKa = 10.03TVTVGPGQAEE236 pKa = 4.18YY237 pKa = 10.67FSTIKK242 pKa = 10.88ASGTATGYY250 pKa = 9.14YY251 pKa = 9.24EE252 pKa = 4.57KK253 pKa = 10.85GVAATTTADD262 pKa = 3.21TTTTEE267 pKa = 4.18EE268 pKa = 4.32TTTTAEE274 pKa = 4.15TTTTAEE280 pKa = 4.1TTTTEE285 pKa = 4.05EE286 pKa = 4.16TTTAEE291 pKa = 4.14TTTTEE296 pKa = 4.11EE297 pKa = 4.29TTTTAEE303 pKa = 4.1TTTEE307 pKa = 4.1FTTEE311 pKa = 3.49TTTYY315 pKa = 7.93FTTEE319 pKa = 3.68TTTDD323 pKa = 3.73FTTEE327 pKa = 3.74TTTDD331 pKa = 3.73FTTEE335 pKa = 3.74TTTDD339 pKa = 3.73FTTEE343 pKa = 3.41TTTYY347 pKa = 9.14VTTDD351 pKa = 3.11TTTVPDD357 pKa = 3.79TTTTMDD363 pKa = 3.25TTTTMDD369 pKa = 3.16TTTAIDD375 pKa = 3.83TTTTTYY381 pKa = 7.38TTTAEE386 pKa = 4.43DD387 pKa = 3.65STTTADD393 pKa = 3.36STTTTDD399 pKa = 5.35LEE401 pKa = 4.65TTTDD405 pKa = 3.24STTTTDD411 pKa = 2.53ITTTADD417 pKa = 3.2TTVPTDD423 pKa = 3.29STAVIDD429 pKa = 3.72STTIIDD435 pKa = 3.79STTTTNPTEE444 pKa = 4.41DD445 pKa = 4.12LSTSIDD451 pKa = 3.47STTTFQSSEE460 pKa = 3.96NTQGSTTDD468 pKa = 3.93DD469 pKa = 4.04GEE471 pKa = 4.79STTTTSDD478 pKa = 3.43LATTDD483 pKa = 3.12TTEE486 pKa = 4.08NLGEE490 pKa = 4.29TTSTAITTSAQIPSIASTEE509 pKa = 4.17SDD511 pKa = 3.35TTTAILSTLEE521 pKa = 3.92SSSALEE527 pKa = 4.06ATPTSQSMTEE537 pKa = 4.01TTPDD541 pKa = 3.32STSDD545 pKa = 3.01ISTLEE550 pKa = 4.08SNEE553 pKa = 4.43PEE555 pKa = 4.3PQPSTASEE563 pKa = 4.52LFTSSVDD570 pKa = 3.48TPTGSVEE577 pKa = 4.13PPSTNTPVGQSSTSALLPEE596 pKa = 4.65TLSPDD601 pKa = 3.12TSSISFPLPEE611 pKa = 4.38SLSTGSDD618 pKa = 3.08SQAPLSTTLSTQDD631 pKa = 3.36SVTVDD636 pKa = 3.62LTASSSPLATSISADD651 pKa = 3.37SSILSSPDD659 pKa = 3.03STSTLSGSTPNASNAPTIEE678 pKa = 3.9RR679 pKa = 11.84YY680 pKa = 10.28VYY682 pKa = 9.78FGCLSSVQGYY692 pKa = 9.1PSFNIVGSATDD703 pKa = 3.7MTPQKK708 pKa = 10.22CISLAAGRR716 pKa = 11.84RR717 pKa = 11.84FVGVHH722 pKa = 5.48LTSCYY727 pKa = 10.83ASDD730 pKa = 3.83SLVSSTLTQDD740 pKa = 3.74ASCDD744 pKa = 3.94LPCPGDD750 pKa = 3.41PTLFCGGFLNADD762 pKa = 3.09GGMSKK767 pKa = 9.31RR768 pKa = 11.84HH769 pKa = 5.02RR770 pKa = 11.84LEE772 pKa = 4.19RR773 pKa = 11.84RR774 pKa = 11.84ISPRR778 pKa = 11.84DD779 pKa = 3.59APPGILLTLYY789 pKa = 10.52LFADD793 pKa = 5.24AIIEE797 pKa = 4.23SSSEE801 pKa = 3.47IAAVSTSSTSPAQTVASQSNSVDD824 pKa = 3.77FSISPTLSEE833 pKa = 4.21PVRR836 pKa = 11.84TSAAGPGDD844 pKa = 3.63VSLSSTLNNPDD855 pKa = 3.26QTSVSAVNSEE865 pKa = 4.59DD866 pKa = 3.26ISLEE870 pKa = 3.88PTLIGPDD877 pKa = 3.34QTSASIGDD885 pKa = 3.74ASLTDD890 pKa = 4.09SPPSVPTTFVPGVNPPPLPSSPFNPGNLSRR920 pKa = 11.84SATFEE925 pKa = 4.67AIVTTVVYY933 pKa = 9.26TVLDD937 pKa = 3.83PHH939 pKa = 7.16DD940 pKa = 4.12PAYY943 pKa = 10.01LTVAEE948 pKa = 4.79FCTTLKK954 pKa = 10.82YY955 pKa = 10.16PPCRR959 pKa = 11.84NCQFQRR965 pKa = 11.84LPTVEE970 pKa = 3.95MTTILTACDD979 pKa = 2.88ACGYY983 pKa = 9.94HH984 pKa = 6.44GEE986 pKa = 4.13NSIEE990 pKa = 3.86LTIPLGANPALMTSNPYY1007 pKa = 9.64TPDD1010 pKa = 3.74DD1011 pKa = 3.97VPGGSPAAAHH1021 pKa = 6.98DD1022 pKa = 4.35EE1023 pKa = 4.31NSGPEE1028 pKa = 3.97YY1029 pKa = 10.39RR1030 pKa = 11.84PRR1032 pKa = 11.84PGGAEE1037 pKa = 3.68ARR1039 pKa = 11.84PTVVPVGHH1047 pKa = 7.09PPYY1050 pKa = 9.94GGHH1053 pKa = 6.84DD1054 pKa = 3.3RR1055 pKa = 11.84PVIHH1059 pKa = 7.2HH1060 pKa = 6.86EE1061 pKa = 4.27GTTGEE1066 pKa = 4.24HH1067 pKa = 5.75NKK1069 pKa = 9.8EE1070 pKa = 4.01NKK1072 pKa = 9.84PPGDD1076 pKa = 3.1NHH1078 pKa = 7.69APIGEE1083 pKa = 4.52PGPSQHH1089 pKa = 5.95EE1090 pKa = 4.09QVPSNHH1096 pKa = 6.6HH1097 pKa = 6.49GEE1099 pKa = 4.28PAGPEE1104 pKa = 3.99YY1105 pKa = 10.44EE1106 pKa = 4.14PQIGPGKK1113 pKa = 10.37DD1114 pKa = 3.1GATVKK1119 pKa = 9.09PTLAPSVEE1127 pKa = 4.22AVPKK1131 pKa = 9.94PEE1133 pKa = 4.11PAGASADD1140 pKa = 3.48LKK1142 pKa = 11.21LFTVPMANIPTTPPDD1157 pKa = 3.4SPVIVAAGHH1166 pKa = 4.75VARR1169 pKa = 11.84GIDD1172 pKa = 3.64GVLTLFALVAGFFLMLL1188 pKa = 4.56
MM1 pKa = 7.64RR2 pKa = 11.84VLLSRR7 pKa = 11.84LIALAIFLPLGVGFTIVTNSDD28 pKa = 3.13GNTLGNAMMPGSGITLVSGTYY49 pKa = 9.28TGASVAAGTFSDD61 pKa = 4.6GPFGISSGTILTSGRR76 pKa = 11.84ADD78 pKa = 3.4GATVAGSDD86 pKa = 4.12NDD88 pKa = 3.78NGQPGYY94 pKa = 10.14MGGNTFDD101 pKa = 4.03AAVLSLDD108 pKa = 3.41ITIAPPFTGFEE119 pKa = 4.32LEE121 pKa = 4.66FAFASNDD128 pKa = 3.38YY129 pKa = 10.93GGQQQFDD136 pKa = 4.38DD137 pKa = 4.17EE138 pKa = 4.83AADD141 pKa = 3.58NTRR144 pKa = 11.84TIFDD148 pKa = 4.14LVNTFMDD155 pKa = 4.3PPYY158 pKa = 10.57SINPPNSLTAYY169 pKa = 9.26DD170 pKa = 4.01KK171 pKa = 11.2SSPPVLIGFATGSGAFNVKK190 pKa = 9.76IAVYY194 pKa = 10.01DD195 pKa = 4.06LNDD198 pKa = 3.64GFEE201 pKa = 4.71DD202 pKa = 3.51SAALIRR208 pKa = 11.84MRR210 pKa = 11.84GCEE213 pKa = 3.82NCAAGPVINYY223 pKa = 5.8EE224 pKa = 4.35TKK226 pKa = 10.03TVTVGPGQAEE236 pKa = 4.18YY237 pKa = 10.67FSTIKK242 pKa = 10.88ASGTATGYY250 pKa = 9.14YY251 pKa = 9.24EE252 pKa = 4.57KK253 pKa = 10.85GVAATTTADD262 pKa = 3.21TTTTEE267 pKa = 4.18EE268 pKa = 4.32TTTTAEE274 pKa = 4.15TTTTAEE280 pKa = 4.1TTTTEE285 pKa = 4.05EE286 pKa = 4.16TTTAEE291 pKa = 4.14TTTTEE296 pKa = 4.11EE297 pKa = 4.29TTTTAEE303 pKa = 4.1TTTEE307 pKa = 4.1FTTEE311 pKa = 3.49TTTYY315 pKa = 7.93FTTEE319 pKa = 3.68TTTDD323 pKa = 3.73FTTEE327 pKa = 3.74TTTDD331 pKa = 3.73FTTEE335 pKa = 3.74TTTDD339 pKa = 3.73FTTEE343 pKa = 3.41TTTYY347 pKa = 9.14VTTDD351 pKa = 3.11TTTVPDD357 pKa = 3.79TTTTMDD363 pKa = 3.25TTTTMDD369 pKa = 3.16TTTAIDD375 pKa = 3.83TTTTTYY381 pKa = 7.38TTTAEE386 pKa = 4.43DD387 pKa = 3.65STTTADD393 pKa = 3.36STTTTDD399 pKa = 5.35LEE401 pKa = 4.65TTTDD405 pKa = 3.24STTTTDD411 pKa = 2.53ITTTADD417 pKa = 3.2TTVPTDD423 pKa = 3.29STAVIDD429 pKa = 3.72STTIIDD435 pKa = 3.79STTTTNPTEE444 pKa = 4.41DD445 pKa = 4.12LSTSIDD451 pKa = 3.47STTTFQSSEE460 pKa = 3.96NTQGSTTDD468 pKa = 3.93DD469 pKa = 4.04GEE471 pKa = 4.79STTTTSDD478 pKa = 3.43LATTDD483 pKa = 3.12TTEE486 pKa = 4.08NLGEE490 pKa = 4.29TTSTAITTSAQIPSIASTEE509 pKa = 4.17SDD511 pKa = 3.35TTTAILSTLEE521 pKa = 3.92SSSALEE527 pKa = 4.06ATPTSQSMTEE537 pKa = 4.01TTPDD541 pKa = 3.32STSDD545 pKa = 3.01ISTLEE550 pKa = 4.08SNEE553 pKa = 4.43PEE555 pKa = 4.3PQPSTASEE563 pKa = 4.52LFTSSVDD570 pKa = 3.48TPTGSVEE577 pKa = 4.13PPSTNTPVGQSSTSALLPEE596 pKa = 4.65TLSPDD601 pKa = 3.12TSSISFPLPEE611 pKa = 4.38SLSTGSDD618 pKa = 3.08SQAPLSTTLSTQDD631 pKa = 3.36SVTVDD636 pKa = 3.62LTASSSPLATSISADD651 pKa = 3.37SSILSSPDD659 pKa = 3.03STSTLSGSTPNASNAPTIEE678 pKa = 3.9RR679 pKa = 11.84YY680 pKa = 10.28VYY682 pKa = 9.78FGCLSSVQGYY692 pKa = 9.1PSFNIVGSATDD703 pKa = 3.7MTPQKK708 pKa = 10.22CISLAAGRR716 pKa = 11.84RR717 pKa = 11.84FVGVHH722 pKa = 5.48LTSCYY727 pKa = 10.83ASDD730 pKa = 3.83SLVSSTLTQDD740 pKa = 3.74ASCDD744 pKa = 3.94LPCPGDD750 pKa = 3.41PTLFCGGFLNADD762 pKa = 3.09GGMSKK767 pKa = 9.31RR768 pKa = 11.84HH769 pKa = 5.02RR770 pKa = 11.84LEE772 pKa = 4.19RR773 pKa = 11.84RR774 pKa = 11.84ISPRR778 pKa = 11.84DD779 pKa = 3.59APPGILLTLYY789 pKa = 10.52LFADD793 pKa = 5.24AIIEE797 pKa = 4.23SSSEE801 pKa = 3.47IAAVSTSSTSPAQTVASQSNSVDD824 pKa = 3.77FSISPTLSEE833 pKa = 4.21PVRR836 pKa = 11.84TSAAGPGDD844 pKa = 3.63VSLSSTLNNPDD855 pKa = 3.26QTSVSAVNSEE865 pKa = 4.59DD866 pKa = 3.26ISLEE870 pKa = 3.88PTLIGPDD877 pKa = 3.34QTSASIGDD885 pKa = 3.74ASLTDD890 pKa = 4.09SPPSVPTTFVPGVNPPPLPSSPFNPGNLSRR920 pKa = 11.84SATFEE925 pKa = 4.67AIVTTVVYY933 pKa = 9.26TVLDD937 pKa = 3.83PHH939 pKa = 7.16DD940 pKa = 4.12PAYY943 pKa = 10.01LTVAEE948 pKa = 4.79FCTTLKK954 pKa = 10.82YY955 pKa = 10.16PPCRR959 pKa = 11.84NCQFQRR965 pKa = 11.84LPTVEE970 pKa = 3.95MTTILTACDD979 pKa = 2.88ACGYY983 pKa = 9.94HH984 pKa = 6.44GEE986 pKa = 4.13NSIEE990 pKa = 3.86LTIPLGANPALMTSNPYY1007 pKa = 9.64TPDD1010 pKa = 3.74DD1011 pKa = 3.97VPGGSPAAAHH1021 pKa = 6.98DD1022 pKa = 4.35EE1023 pKa = 4.31NSGPEE1028 pKa = 3.97YY1029 pKa = 10.39RR1030 pKa = 11.84PRR1032 pKa = 11.84PGGAEE1037 pKa = 3.68ARR1039 pKa = 11.84PTVVPVGHH1047 pKa = 7.09PPYY1050 pKa = 9.94GGHH1053 pKa = 6.84DD1054 pKa = 3.3RR1055 pKa = 11.84PVIHH1059 pKa = 7.2HH1060 pKa = 6.86EE1061 pKa = 4.27GTTGEE1066 pKa = 4.24HH1067 pKa = 5.75NKK1069 pKa = 9.8EE1070 pKa = 4.01NKK1072 pKa = 9.84PPGDD1076 pKa = 3.1NHH1078 pKa = 7.69APIGEE1083 pKa = 4.52PGPSQHH1089 pKa = 5.95EE1090 pKa = 4.09QVPSNHH1096 pKa = 6.6HH1097 pKa = 6.49GEE1099 pKa = 4.28PAGPEE1104 pKa = 3.99YY1105 pKa = 10.44EE1106 pKa = 4.14PQIGPGKK1113 pKa = 10.37DD1114 pKa = 3.1GATVKK1119 pKa = 9.09PTLAPSVEE1127 pKa = 4.22AVPKK1131 pKa = 9.94PEE1133 pKa = 4.11PAGASADD1140 pKa = 3.48LKK1142 pKa = 11.21LFTVPMANIPTTPPDD1157 pKa = 3.4SPVIVAAGHH1166 pKa = 4.75VARR1169 pKa = 11.84GIDD1172 pKa = 3.64GVLTLFALVAGFFLMLL1188 pKa = 4.56
Molecular weight: 122.89 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C7YRP9|C7YRP9_FUSV7 Uncharacterized protein CHR2115 OS=Fusarium vanettenii (strain ATCC MYA-4622 / CBS 123669 / FGSC 9596 / NRRL 45880 / 77-13-4) OX=660122 GN=CHR2115 PE=3 SV=1
MM1 pKa = 7.86PLTRR5 pKa = 11.84THH7 pKa = 6.65RR8 pKa = 11.84QTTPRR13 pKa = 11.84RR14 pKa = 11.84SIFSTRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84APARR26 pKa = 11.84SHH28 pKa = 5.86HH29 pKa = 6.11TVTTTTTTTRR39 pKa = 11.84KK40 pKa = 8.7PRR42 pKa = 11.84RR43 pKa = 11.84GFLGGGRR50 pKa = 11.84RR51 pKa = 11.84THH53 pKa = 7.04AAPVHH58 pKa = 5.1HH59 pKa = 5.69QQRR62 pKa = 11.84RR63 pKa = 11.84PSMKK67 pKa = 10.24DD68 pKa = 2.95KK69 pKa = 11.37VSGALLRR76 pKa = 11.84LKK78 pKa = 10.81GSLTRR83 pKa = 11.84RR84 pKa = 11.84PGVKK88 pKa = 9.89AAGTRR93 pKa = 11.84RR94 pKa = 11.84MHH96 pKa = 5.68GTDD99 pKa = 2.6GRR101 pKa = 11.84GSHH104 pKa = 5.81RR105 pKa = 11.84RR106 pKa = 11.84ARR108 pKa = 11.84HH109 pKa = 4.06FF110 pKa = 3.77
MM1 pKa = 7.86PLTRR5 pKa = 11.84THH7 pKa = 6.65RR8 pKa = 11.84QTTPRR13 pKa = 11.84RR14 pKa = 11.84SIFSTRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84APARR26 pKa = 11.84SHH28 pKa = 5.86HH29 pKa = 6.11TVTTTTTTTRR39 pKa = 11.84KK40 pKa = 8.7PRR42 pKa = 11.84RR43 pKa = 11.84GFLGGGRR50 pKa = 11.84RR51 pKa = 11.84THH53 pKa = 7.04AAPVHH58 pKa = 5.1HH59 pKa = 5.69QQRR62 pKa = 11.84RR63 pKa = 11.84PSMKK67 pKa = 10.24DD68 pKa = 2.95KK69 pKa = 11.37VSGALLRR76 pKa = 11.84LKK78 pKa = 10.81GSLTRR83 pKa = 11.84RR84 pKa = 11.84PGVKK88 pKa = 9.89AAGTRR93 pKa = 11.84RR94 pKa = 11.84MHH96 pKa = 5.68GTDD99 pKa = 2.6GRR101 pKa = 11.84GSHH104 pKa = 5.81RR105 pKa = 11.84RR106 pKa = 11.84ARR108 pKa = 11.84HH109 pKa = 4.06FF110 pKa = 3.77
Molecular weight: 12.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7536017 |
48 |
7649 |
479.7 |
53.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.48 ± 0.02 | 1.354 ± 0.008 |
5.815 ± 0.015 | 6.178 ± 0.019 |
3.891 ± 0.012 | 6.942 ± 0.017 |
2.392 ± 0.01 | 5.029 ± 0.011 |
4.833 ± 0.016 | 9.045 ± 0.02 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.225 ± 0.007 | 3.589 ± 0.01 |
5.943 ± 0.018 | 3.894 ± 0.013 |
5.958 ± 0.017 | 7.865 ± 0.019 |
5.882 ± 0.016 | 6.235 ± 0.014 |
1.647 ± 0.007 | 2.803 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
