
Beihai tombus-like virus 12
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.22
Get precalculated fractions of proteins
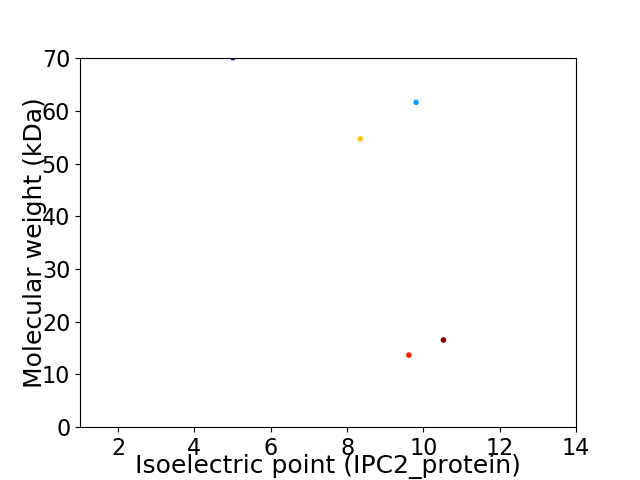
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFF1|A0A1L3KFF1_9VIRU Uncharacterized protein OS=Beihai tombus-like virus 12 OX=1922715 PE=4 SV=1
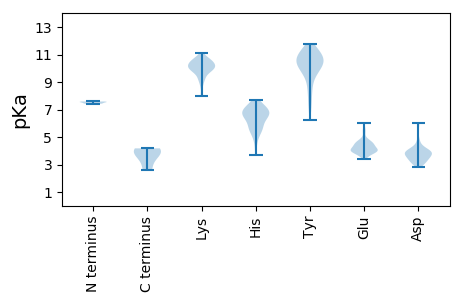
MM1 pKa = 7.56AGRR4 pKa = 11.84RR5 pKa = 11.84GRR7 pKa = 11.84QRR9 pKa = 11.84GATRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84GNALASAQRR24 pKa = 11.84VLHH27 pKa = 5.6RR28 pKa = 11.84TSIPFDD34 pKa = 3.4VADD37 pKa = 3.96ATGASCTKK45 pKa = 10.24IVLDD49 pKa = 3.96PSAYY53 pKa = 8.99TKK55 pKa = 10.91DD56 pKa = 3.42RR57 pKa = 11.84ASAMASIFQKK67 pKa = 10.06YY68 pKa = 8.38AVRR71 pKa = 11.84SVAVDD76 pKa = 3.86FTPSVGFNEE85 pKa = 5.54SSGQWGVGYY94 pKa = 8.55MADD97 pKa = 4.68EE98 pKa = 4.63SNQGQGLPSTTADD111 pKa = 3.28VVAMARR117 pKa = 11.84QGCGTLSPVRR127 pKa = 11.84SKK129 pKa = 10.72LHH131 pKa = 4.95WAVPRR136 pKa = 11.84RR137 pKa = 11.84SLRR140 pKa = 11.84NITEE144 pKa = 4.78GYY146 pKa = 7.72TPGGAEE152 pKa = 3.56QPGAVFVGVAAAASVAQPGVWTLDD176 pKa = 2.94ILYY179 pKa = 8.4EE180 pKa = 4.16FWGPRR185 pKa = 11.84PPFEE189 pKa = 4.24KK190 pKa = 10.5AVLPTEE196 pKa = 4.05ITSGGGYY203 pKa = 10.44GITFGDD209 pKa = 4.27GSDD212 pKa = 3.21WPYY215 pKa = 11.24GYY217 pKa = 10.37HH218 pKa = 6.98PPDD221 pKa = 3.89ALWRR225 pKa = 11.84NLWRR229 pKa = 11.84ARR231 pKa = 11.84DD232 pKa = 3.8SCSLTTVTSFYY243 pKa = 10.71AHH245 pKa = 6.61TSAGVFLLEE254 pKa = 4.39GPGKK258 pKa = 9.8IEE260 pKa = 3.73QLEE263 pKa = 4.14GLGTVEE269 pKa = 4.76VINAFMDD276 pKa = 3.54THH278 pKa = 6.94SLLGVSLFGTNAMKK292 pKa = 10.21QIPIPAVRR300 pKa = 11.84QAVSVQPEE308 pKa = 3.64FRR310 pKa = 11.84VRR312 pKa = 11.84VINEE316 pKa = 3.39VDD318 pKa = 3.93DD319 pKa = 4.22PVPTQEE325 pKa = 4.61TVDD328 pKa = 3.95PLPVEE333 pKa = 4.47VTNVPLPVAEE343 pKa = 4.48TVDD346 pKa = 3.91PLPVEE351 pKa = 4.47VTNVPLPVAEE361 pKa = 4.48TVDD364 pKa = 3.91PLPVEE369 pKa = 4.47VTNVPLPVAEE379 pKa = 4.48TVDD382 pKa = 3.91PLPVEE387 pKa = 4.47VTNVPLPVAEE397 pKa = 4.48TVDD400 pKa = 4.05PLPVDD405 pKa = 3.62VTNTPLPVAEE415 pKa = 4.68VVDD418 pKa = 4.19PLPVKK423 pKa = 9.29VTNTPLPVAEE433 pKa = 4.72AQLPLPVNEE442 pKa = 4.7TNLPLPVDD450 pKa = 4.1DD451 pKa = 4.89GGGGFFMNLATGVITAVVKK470 pKa = 10.15QAGRR474 pKa = 11.84TYY476 pKa = 10.56TRR478 pKa = 11.84DD479 pKa = 3.49SKK481 pKa = 9.82EE482 pKa = 4.0TPAGYY487 pKa = 10.36VRR489 pKa = 11.84LKK491 pKa = 10.45CSYY494 pKa = 9.34PVGGAVDD501 pKa = 3.93GQVYY505 pKa = 10.61GSVNIPAYY513 pKa = 7.1PTAEE517 pKa = 3.96AGLTHH522 pKa = 7.41FGRR525 pKa = 11.84LDD527 pKa = 3.14AGYY530 pKa = 10.07DD531 pKa = 3.6YY532 pKa = 11.42ASEE535 pKa = 4.59LFWKK539 pKa = 10.13RR540 pKa = 11.84FANMTVGLPNDD551 pKa = 3.14HH552 pKa = 7.06WYY554 pKa = 10.58KK555 pKa = 10.66AVSSEE560 pKa = 3.96PPPFGGAGGMYY571 pKa = 9.7IYY573 pKa = 9.66ITDD576 pKa = 3.47TRR578 pKa = 11.84KK579 pKa = 9.36PAGLFAGNHH588 pKa = 3.68ITARR592 pKa = 11.84CRR594 pKa = 11.84WTSAEE599 pKa = 3.86WFEE602 pKa = 4.0VHH604 pKa = 6.49CWHH607 pKa = 6.75NVDD610 pKa = 6.0SDD612 pKa = 4.16ASWSKK617 pKa = 10.9DD618 pKa = 3.2VHH620 pKa = 6.61CFLLHH625 pKa = 6.29KK626 pKa = 10.01PLYY629 pKa = 10.37DD630 pKa = 3.84SEE632 pKa = 5.99LDD634 pKa = 3.85DD635 pKa = 6.05VYY637 pKa = 11.21DD638 pKa = 3.88TYY640 pKa = 11.41QDD642 pKa = 3.21HH643 pKa = 6.92RR644 pKa = 11.84VRR646 pKa = 11.84THH648 pKa = 6.28PVV650 pKa = 2.58
MM1 pKa = 7.56AGRR4 pKa = 11.84RR5 pKa = 11.84GRR7 pKa = 11.84QRR9 pKa = 11.84GATRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84GNALASAQRR24 pKa = 11.84VLHH27 pKa = 5.6RR28 pKa = 11.84TSIPFDD34 pKa = 3.4VADD37 pKa = 3.96ATGASCTKK45 pKa = 10.24IVLDD49 pKa = 3.96PSAYY53 pKa = 8.99TKK55 pKa = 10.91DD56 pKa = 3.42RR57 pKa = 11.84ASAMASIFQKK67 pKa = 10.06YY68 pKa = 8.38AVRR71 pKa = 11.84SVAVDD76 pKa = 3.86FTPSVGFNEE85 pKa = 5.54SSGQWGVGYY94 pKa = 8.55MADD97 pKa = 4.68EE98 pKa = 4.63SNQGQGLPSTTADD111 pKa = 3.28VVAMARR117 pKa = 11.84QGCGTLSPVRR127 pKa = 11.84SKK129 pKa = 10.72LHH131 pKa = 4.95WAVPRR136 pKa = 11.84RR137 pKa = 11.84SLRR140 pKa = 11.84NITEE144 pKa = 4.78GYY146 pKa = 7.72TPGGAEE152 pKa = 3.56QPGAVFVGVAAAASVAQPGVWTLDD176 pKa = 2.94ILYY179 pKa = 8.4EE180 pKa = 4.16FWGPRR185 pKa = 11.84PPFEE189 pKa = 4.24KK190 pKa = 10.5AVLPTEE196 pKa = 4.05ITSGGGYY203 pKa = 10.44GITFGDD209 pKa = 4.27GSDD212 pKa = 3.21WPYY215 pKa = 11.24GYY217 pKa = 10.37HH218 pKa = 6.98PPDD221 pKa = 3.89ALWRR225 pKa = 11.84NLWRR229 pKa = 11.84ARR231 pKa = 11.84DD232 pKa = 3.8SCSLTTVTSFYY243 pKa = 10.71AHH245 pKa = 6.61TSAGVFLLEE254 pKa = 4.39GPGKK258 pKa = 9.8IEE260 pKa = 3.73QLEE263 pKa = 4.14GLGTVEE269 pKa = 4.76VINAFMDD276 pKa = 3.54THH278 pKa = 6.94SLLGVSLFGTNAMKK292 pKa = 10.21QIPIPAVRR300 pKa = 11.84QAVSVQPEE308 pKa = 3.64FRR310 pKa = 11.84VRR312 pKa = 11.84VINEE316 pKa = 3.39VDD318 pKa = 3.93DD319 pKa = 4.22PVPTQEE325 pKa = 4.61TVDD328 pKa = 3.95PLPVEE333 pKa = 4.47VTNVPLPVAEE343 pKa = 4.48TVDD346 pKa = 3.91PLPVEE351 pKa = 4.47VTNVPLPVAEE361 pKa = 4.48TVDD364 pKa = 3.91PLPVEE369 pKa = 4.47VTNVPLPVAEE379 pKa = 4.48TVDD382 pKa = 3.91PLPVEE387 pKa = 4.47VTNVPLPVAEE397 pKa = 4.48TVDD400 pKa = 4.05PLPVDD405 pKa = 3.62VTNTPLPVAEE415 pKa = 4.68VVDD418 pKa = 4.19PLPVKK423 pKa = 9.29VTNTPLPVAEE433 pKa = 4.72AQLPLPVNEE442 pKa = 4.7TNLPLPVDD450 pKa = 4.1DD451 pKa = 4.89GGGGFFMNLATGVITAVVKK470 pKa = 10.15QAGRR474 pKa = 11.84TYY476 pKa = 10.56TRR478 pKa = 11.84DD479 pKa = 3.49SKK481 pKa = 9.82EE482 pKa = 4.0TPAGYY487 pKa = 10.36VRR489 pKa = 11.84LKK491 pKa = 10.45CSYY494 pKa = 9.34PVGGAVDD501 pKa = 3.93GQVYY505 pKa = 10.61GSVNIPAYY513 pKa = 7.1PTAEE517 pKa = 3.96AGLTHH522 pKa = 7.41FGRR525 pKa = 11.84LDD527 pKa = 3.14AGYY530 pKa = 10.07DD531 pKa = 3.6YY532 pKa = 11.42ASEE535 pKa = 4.59LFWKK539 pKa = 10.13RR540 pKa = 11.84FANMTVGLPNDD551 pKa = 3.14HH552 pKa = 7.06WYY554 pKa = 10.58KK555 pKa = 10.66AVSSEE560 pKa = 3.96PPPFGGAGGMYY571 pKa = 9.7IYY573 pKa = 9.66ITDD576 pKa = 3.47TRR578 pKa = 11.84KK579 pKa = 9.36PAGLFAGNHH588 pKa = 3.68ITARR592 pKa = 11.84CRR594 pKa = 11.84WTSAEE599 pKa = 3.86WFEE602 pKa = 4.0VHH604 pKa = 6.49CWHH607 pKa = 6.75NVDD610 pKa = 6.0SDD612 pKa = 4.16ASWSKK617 pKa = 10.9DD618 pKa = 3.2VHH620 pKa = 6.61CFLLHH625 pKa = 6.29KK626 pKa = 10.01PLYY629 pKa = 10.37DD630 pKa = 3.84SEE632 pKa = 5.99LDD634 pKa = 3.85DD635 pKa = 6.05VYY637 pKa = 11.21DD638 pKa = 3.88TYY640 pKa = 11.41QDD642 pKa = 3.21HH643 pKa = 6.92RR644 pKa = 11.84VRR646 pKa = 11.84THH648 pKa = 6.28PVV650 pKa = 2.58
Molecular weight: 70.11 kDa
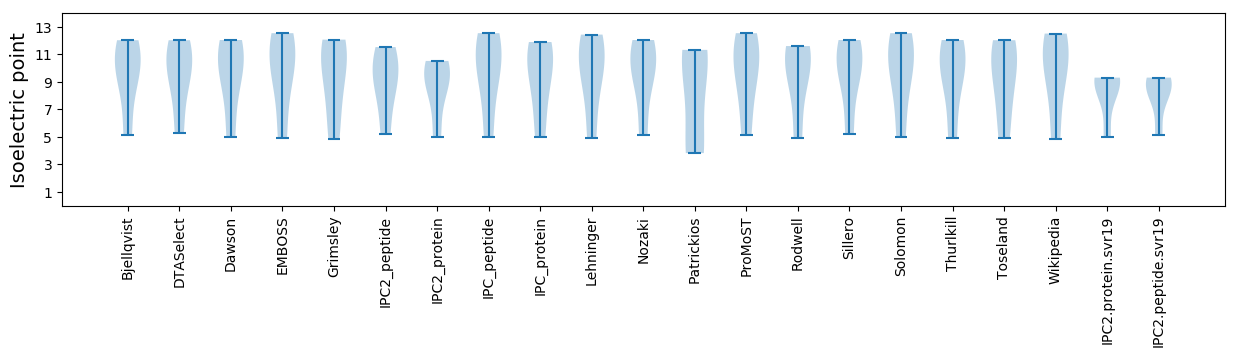
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFE5|A0A1L3KFE5_9VIRU Uncharacterized protein OS=Beihai tombus-like virus 12 OX=1922715 PE=4 SV=1
MM1 pKa = 7.42GAAQTVVRR9 pKa = 11.84RR10 pKa = 11.84DD11 pKa = 3.41YY12 pKa = 11.36AGVDD16 pKa = 4.01FGHH19 pKa = 7.7AARR22 pKa = 11.84STAVVPIVVIIGLVSVPLLIYY43 pKa = 10.3FRR45 pKa = 11.84SSGRR49 pKa = 11.84WCLGALQGLLRR60 pKa = 11.84SARR63 pKa = 11.84ASWLGSKK70 pKa = 8.68YY71 pKa = 9.64HH72 pKa = 5.74QRR74 pKa = 11.84LVVPARR80 pKa = 11.84IMFQDD85 pKa = 3.04AHH87 pKa = 7.27DD88 pKa = 4.46GVLDD92 pKa = 3.53VAARR96 pKa = 11.84KK97 pKa = 9.67LGGMIANRR105 pKa = 11.84ARR107 pKa = 11.84FYY109 pKa = 11.45EE110 pKa = 4.23EE111 pKa = 5.28LDD113 pKa = 3.58VPLLMIHH120 pKa = 5.43EE121 pKa = 4.46QVRR124 pKa = 3.52
MM1 pKa = 7.42GAAQTVVRR9 pKa = 11.84RR10 pKa = 11.84DD11 pKa = 3.41YY12 pKa = 11.36AGVDD16 pKa = 4.01FGHH19 pKa = 7.7AARR22 pKa = 11.84STAVVPIVVIIGLVSVPLLIYY43 pKa = 10.3FRR45 pKa = 11.84SSGRR49 pKa = 11.84WCLGALQGLLRR60 pKa = 11.84SARR63 pKa = 11.84ASWLGSKK70 pKa = 8.68YY71 pKa = 9.64HH72 pKa = 5.74QRR74 pKa = 11.84LVVPARR80 pKa = 11.84IMFQDD85 pKa = 3.04AHH87 pKa = 7.27DD88 pKa = 4.46GVLDD92 pKa = 3.53VAARR96 pKa = 11.84KK97 pKa = 9.67LGGMIANRR105 pKa = 11.84ARR107 pKa = 11.84FYY109 pKa = 11.45EE110 pKa = 4.23EE111 pKa = 5.28LDD113 pKa = 3.58VPLLMIHH120 pKa = 5.43EE121 pKa = 4.46QVRR124 pKa = 3.52
Molecular weight: 13.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1977 |
124 |
650 |
395.4 |
43.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.521 ± 0.532 | 2.175 ± 0.523 |
4.148 ± 0.711 | 4.451 ± 0.514 |
2.833 ± 0.758 | 7.688 ± 0.683 |
1.973 ± 0.377 | 2.479 ± 0.483 |
3.794 ± 1.026 | 7.436 ± 1.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.074 ± 0.498 | 2.681 ± 0.36 |
8.346 ± 1.148 | 3.44 ± 0.696 |
9.256 ± 1.624 | 6.677 ± 0.655 |
6.626 ± 1.261 | 8.7 ± 1.324 |
1.669 ± 0.273 | 3.035 ± 0.857 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
