
Chloroherpeton thalassium (strain ATCC 35110 / GB-78)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Chlorobi; Chlorobia; Chlorobiales; Chlorobiaceae; Chloroherpeton; Chloroherpeton thalassium
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
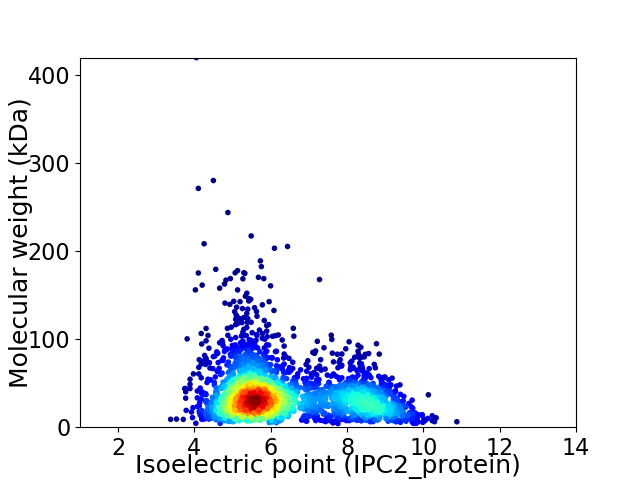
Virtual 2D-PAGE plot for 2708 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
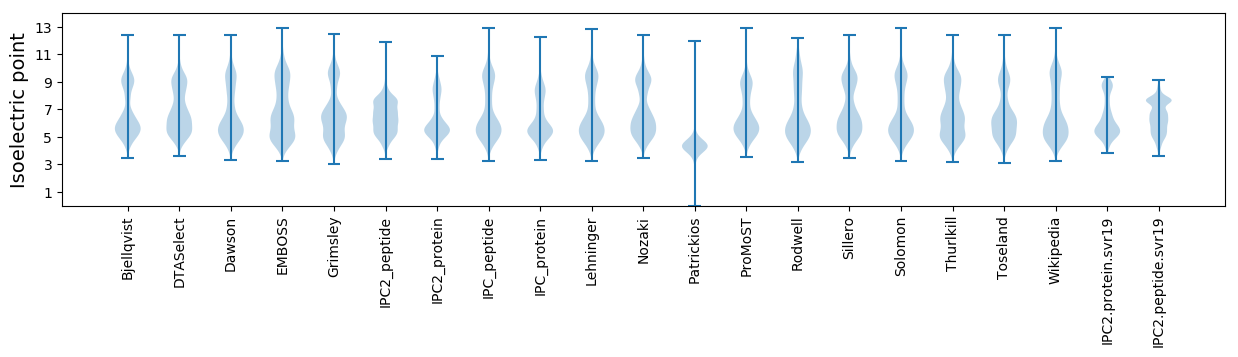
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B3QVI0|B3QVI0_CHLT3 Metallophosphoesterase OS=Chloroherpeton thalassium (strain ATCC 35110 / GB-78) OX=517418 GN=Ctha_2128 PE=3 SV=1
MM1 pKa = 7.72RR2 pKa = 11.84EE3 pKa = 4.57KK4 pKa = 10.9SITWKK9 pKa = 10.59NAVLKK14 pKa = 10.63VLSLLLIFAFAGCDD28 pKa = 4.13DD29 pKa = 5.39DD30 pKa = 5.94DD31 pKa = 6.21CPTQEE36 pKa = 5.46CSTTTCAGADD46 pKa = 3.34AALVLSEE53 pKa = 5.38GNYY56 pKa = 9.95EE57 pKa = 4.58SNNSTLTHH65 pKa = 6.68YY66 pKa = 11.34DD67 pKa = 2.97MEE69 pKa = 5.15TNVATQNYY77 pKa = 7.76FEE79 pKa = 4.02QRR81 pKa = 11.84IGRR84 pKa = 11.84LLGDD88 pKa = 4.14TGNDD92 pKa = 3.3MILVDD97 pKa = 4.64NILYY101 pKa = 10.17IVVNNSNKK109 pKa = 10.35LEE111 pKa = 4.12VVDD114 pKa = 3.81TDD116 pKa = 3.1TWTSLAKK123 pKa = 9.11ITLEE127 pKa = 3.95KK128 pKa = 10.76DD129 pKa = 3.19EE130 pKa = 5.2SGSSPRR136 pKa = 11.84EE137 pKa = 3.45IVEE140 pKa = 4.06YY141 pKa = 10.42NGKK144 pKa = 9.68LFISNFNGYY153 pKa = 8.8VAVVDD158 pKa = 3.95TANSYY163 pKa = 11.1GNEE166 pKa = 4.01IEE168 pKa = 4.27WIQVGTQPNGITVLDD183 pKa = 3.97GKK185 pKa = 10.8IYY187 pKa = 10.34VANSNYY193 pKa = 10.55DD194 pKa = 3.03NGYY197 pKa = 8.55NPGSISVIDD206 pKa = 3.9PTTLTVTQTLSDD218 pKa = 3.07IGVNISSIATDD229 pKa = 4.1DD230 pKa = 3.82YY231 pKa = 11.44GDD233 pKa = 4.56LYY235 pKa = 11.36VISKK239 pKa = 10.68GNYY242 pKa = 8.32SDD244 pKa = 5.26ASANLYY250 pKa = 8.99VVNPTTGAITKK261 pKa = 8.73TFDD264 pKa = 2.63ITAQSIAIHH273 pKa = 6.52GDD275 pKa = 2.92VGYY278 pKa = 10.95VSVGGYY284 pKa = 8.97DD285 pKa = 4.02ANWNYY290 pKa = 9.93VASIYY295 pKa = 10.69KK296 pKa = 9.7IDD298 pKa = 3.83VEE300 pKa = 4.67ADD302 pKa = 3.14TVMTEE307 pKa = 4.71DD308 pKa = 5.24FIEE311 pKa = 4.38SSNFQTLYY319 pKa = 10.99GLNVDD324 pKa = 4.6PNTGDD329 pKa = 3.55IYY331 pKa = 11.4CLDD334 pKa = 3.91ALDD337 pKa = 4.31YY338 pKa = 10.66SVSGTVTVFDD348 pKa = 4.68EE349 pKa = 5.77DD350 pKa = 4.16GQQTTSFKK358 pKa = 11.16SGVNPCKK365 pKa = 10.73VLFLEE370 pKa = 5.13DD371 pKa = 3.16
MM1 pKa = 7.72RR2 pKa = 11.84EE3 pKa = 4.57KK4 pKa = 10.9SITWKK9 pKa = 10.59NAVLKK14 pKa = 10.63VLSLLLIFAFAGCDD28 pKa = 4.13DD29 pKa = 5.39DD30 pKa = 5.94DD31 pKa = 6.21CPTQEE36 pKa = 5.46CSTTTCAGADD46 pKa = 3.34AALVLSEE53 pKa = 5.38GNYY56 pKa = 9.95EE57 pKa = 4.58SNNSTLTHH65 pKa = 6.68YY66 pKa = 11.34DD67 pKa = 2.97MEE69 pKa = 5.15TNVATQNYY77 pKa = 7.76FEE79 pKa = 4.02QRR81 pKa = 11.84IGRR84 pKa = 11.84LLGDD88 pKa = 4.14TGNDD92 pKa = 3.3MILVDD97 pKa = 4.64NILYY101 pKa = 10.17IVVNNSNKK109 pKa = 10.35LEE111 pKa = 4.12VVDD114 pKa = 3.81TDD116 pKa = 3.1TWTSLAKK123 pKa = 9.11ITLEE127 pKa = 3.95KK128 pKa = 10.76DD129 pKa = 3.19EE130 pKa = 5.2SGSSPRR136 pKa = 11.84EE137 pKa = 3.45IVEE140 pKa = 4.06YY141 pKa = 10.42NGKK144 pKa = 9.68LFISNFNGYY153 pKa = 8.8VAVVDD158 pKa = 3.95TANSYY163 pKa = 11.1GNEE166 pKa = 4.01IEE168 pKa = 4.27WIQVGTQPNGITVLDD183 pKa = 3.97GKK185 pKa = 10.8IYY187 pKa = 10.34VANSNYY193 pKa = 10.55DD194 pKa = 3.03NGYY197 pKa = 8.55NPGSISVIDD206 pKa = 3.9PTTLTVTQTLSDD218 pKa = 3.07IGVNISSIATDD229 pKa = 4.1DD230 pKa = 3.82YY231 pKa = 11.44GDD233 pKa = 4.56LYY235 pKa = 11.36VISKK239 pKa = 10.68GNYY242 pKa = 8.32SDD244 pKa = 5.26ASANLYY250 pKa = 8.99VVNPTTGAITKK261 pKa = 8.73TFDD264 pKa = 2.63ITAQSIAIHH273 pKa = 6.52GDD275 pKa = 2.92VGYY278 pKa = 10.95VSVGGYY284 pKa = 8.97DD285 pKa = 4.02ANWNYY290 pKa = 9.93VASIYY295 pKa = 10.69KK296 pKa = 9.7IDD298 pKa = 3.83VEE300 pKa = 4.67ADD302 pKa = 3.14TVMTEE307 pKa = 4.71DD308 pKa = 5.24FIEE311 pKa = 4.38SSNFQTLYY319 pKa = 10.99GLNVDD324 pKa = 4.6PNTGDD329 pKa = 3.55IYY331 pKa = 11.4CLDD334 pKa = 3.91ALDD337 pKa = 4.31YY338 pKa = 10.66SVSGTVTVFDD348 pKa = 4.68EE349 pKa = 5.77DD350 pKa = 4.16GQQTTSFKK358 pKa = 11.16SGVNPCKK365 pKa = 10.73VLFLEE370 pKa = 5.13DD371 pKa = 3.16
Molecular weight: 40.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B3QYW1|B3QYW1_CHLT3 Redox-active disulfide protein 2 OS=Chloroherpeton thalassium (strain ATCC 35110 / GB-78) OX=517418 GN=Ctha_1190 PE=4 SV=1
MM1 pKa = 7.28KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.06QPHH8 pKa = 4.63NRR10 pKa = 11.84KK11 pKa = 9.12RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.06HH16 pKa = 3.97GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 9.25VLSARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.11GRR39 pKa = 11.84HH40 pKa = 5.33RR41 pKa = 11.84LTVSSEE47 pKa = 4.03TSNKK51 pKa = 9.41KK52 pKa = 10.03
MM1 pKa = 7.28KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.06QPHH8 pKa = 4.63NRR10 pKa = 11.84KK11 pKa = 9.12RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.06HH16 pKa = 3.97GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 9.25VLSARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.11GRR39 pKa = 11.84HH40 pKa = 5.33RR41 pKa = 11.84LTVSSEE47 pKa = 4.03TSNKK51 pKa = 9.41KK52 pKa = 10.03
Molecular weight: 6.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
947848 |
36 |
3944 |
350.0 |
39.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.114 ± 0.047 | 0.898 ± 0.017 |
5.218 ± 0.039 | 7.159 ± 0.05 |
4.958 ± 0.033 | 6.62 ± 0.045 |
2.031 ± 0.024 | 6.889 ± 0.042 |
6.569 ± 0.049 | 9.93 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.33 ± 0.022 | 4.306 ± 0.033 |
3.82 ± 0.027 | 3.602 ± 0.029 |
4.505 ± 0.035 | 6.813 ± 0.051 |
5.272 ± 0.045 | 6.509 ± 0.035 |
0.983 ± 0.017 | 3.471 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
