
Deinococcus deserti (strain DSM 17065 / CIP 109153 / LMG 22923 / VCD115)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Deinococcus-Thermus; Deinococci; Deinococcales; Deinococcaceae; Deinococcus; Deinococcus deserti
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
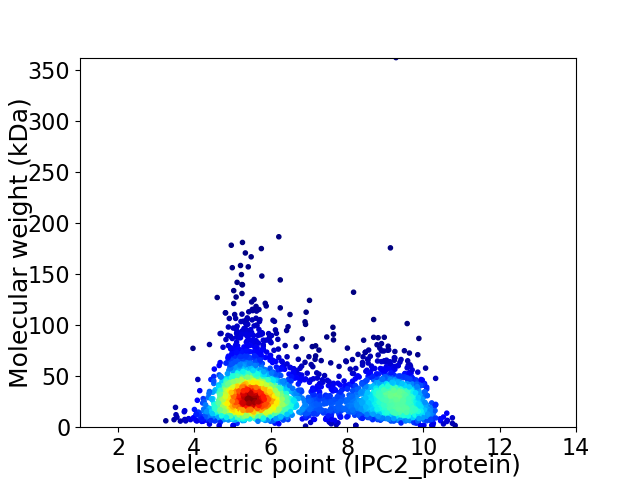
Virtual 2D-PAGE plot for 3459 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C1D3P1|C1D3P1_DEIDV Uncharacterized protein OS=Deinococcus deserti (strain DSM 17065 / CIP 109153 / LMG 22923 / VCD115) OX=546414 GN=Deide_3p01740 PE=4 SV=1
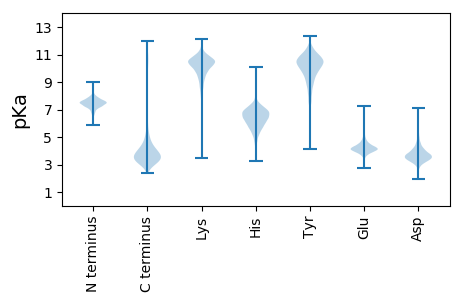
MM1 pKa = 7.87AKK3 pKa = 8.89GTLDD7 pKa = 3.2TSRR10 pKa = 11.84IRR12 pKa = 11.84DD13 pKa = 3.33WEE15 pKa = 4.35TFHH18 pKa = 7.15DD19 pKa = 3.87VSMQAFGFPDD29 pKa = 4.69FYY31 pKa = 11.12GRR33 pKa = 11.84NMDD36 pKa = 3.52AWIDD40 pKa = 3.66CLTYY44 pKa = 10.6LDD46 pKa = 4.81EE47 pKa = 6.3ADD49 pKa = 3.85GMSSVVLGSDD59 pKa = 3.55EE60 pKa = 4.57LLCIHH65 pKa = 6.31VPGFSIFAASLPDD78 pKa = 3.37VANAFLACSCFHH90 pKa = 6.02QPAVYY95 pKa = 8.5RR96 pKa = 11.84TEE98 pKa = 4.27GCFPTDD104 pKa = 3.58FDD106 pKa = 4.14AA107 pKa = 6.82
MM1 pKa = 7.87AKK3 pKa = 8.89GTLDD7 pKa = 3.2TSRR10 pKa = 11.84IRR12 pKa = 11.84DD13 pKa = 3.33WEE15 pKa = 4.35TFHH18 pKa = 7.15DD19 pKa = 3.87VSMQAFGFPDD29 pKa = 4.69FYY31 pKa = 11.12GRR33 pKa = 11.84NMDD36 pKa = 3.52AWIDD40 pKa = 3.66CLTYY44 pKa = 10.6LDD46 pKa = 4.81EE47 pKa = 6.3ADD49 pKa = 3.85GMSSVVLGSDD59 pKa = 3.55EE60 pKa = 4.57LLCIHH65 pKa = 6.31VPGFSIFAASLPDD78 pKa = 3.37VANAFLACSCFHH90 pKa = 6.02QPAVYY95 pKa = 8.5RR96 pKa = 11.84TEE98 pKa = 4.27GCFPTDD104 pKa = 3.58FDD106 pKa = 4.14AA107 pKa = 6.82
Molecular weight: 11.86 kDa
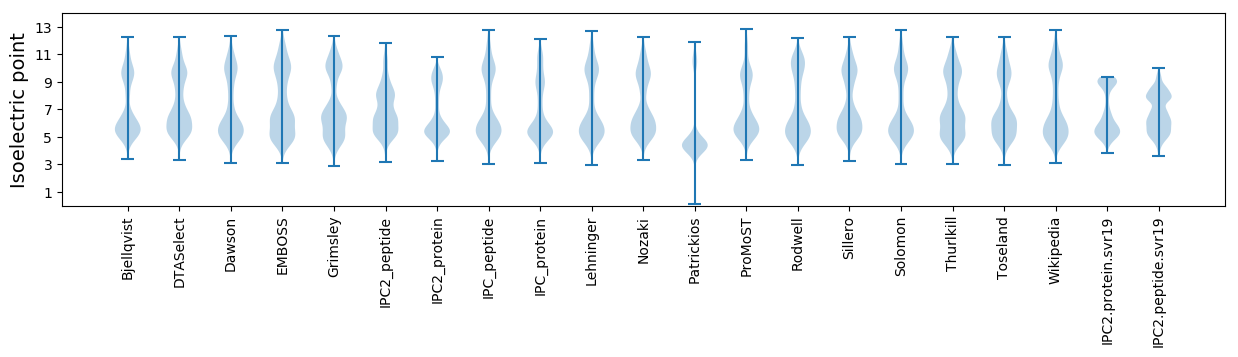
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C1CV97|C1CV97_DEIDV Putative DegV family protein OS=Deinococcus deserti (strain DSM 17065 / CIP 109153 / LMG 22923 / VCD115) OX=546414 GN=Deide_12040 PE=4 SV=1
MM1 pKa = 8.12DD2 pKa = 4.81SLGALAAQFGIANLFGRR19 pKa = 11.84GFISVIITFFAAWVFTWQFIPRR41 pKa = 11.84LRR43 pKa = 11.84DD44 pKa = 3.26FAVKK48 pKa = 10.32AGWADD53 pKa = 3.35QPNARR58 pKa = 11.84RR59 pKa = 11.84LNKK62 pKa = 10.16EE63 pKa = 3.86PLPNAGGLAIFAGFITSVVLAWALRR88 pKa = 11.84PIMVEE93 pKa = 3.66LVNIQVLSILLGASIMVLVGFIDD116 pKa = 3.86DD117 pKa = 3.94QFGLSPVTRR126 pKa = 11.84LVVQVLVAVLLVVNGLKK143 pKa = 9.43MDD145 pKa = 4.5FNAIPFLPVLPEE157 pKa = 3.99AVNEE161 pKa = 4.05PLSTLLTIVWIVGLTNAVNLMDD183 pKa = 4.56GVDD186 pKa = 3.82GVVGGVGFVVSMVLLVTAAQFTDD209 pKa = 3.37RR210 pKa = 11.84AAAVILLAGLAGATLGYY227 pKa = 10.07LRR229 pKa = 11.84HH230 pKa = 6.18NFNPSRR236 pKa = 11.84IIMGDD241 pKa = 2.77AGAYY245 pKa = 10.48LIGFTLAAVSLLGTLKK261 pKa = 10.5FSAGASLIVPLIVLALPVLDD281 pKa = 3.76TTQVVIGRR289 pKa = 11.84LARR292 pKa = 11.84GIRR295 pKa = 11.84NPLGHH300 pKa = 7.52PDD302 pKa = 3.16KK303 pKa = 10.5THH305 pKa = 4.92IHH307 pKa = 5.91HH308 pKa = 6.94RR309 pKa = 11.84VLARR313 pKa = 11.84TASARR318 pKa = 11.84RR319 pKa = 11.84TAVILWMVALACGMLGMLLQGVPLLAIGLTGVLVLVCLWFVAHH362 pKa = 6.21RR363 pKa = 11.84RR364 pKa = 11.84VRR366 pKa = 11.84AHH368 pKa = 5.87SRR370 pKa = 11.84EE371 pKa = 4.12DD372 pKa = 3.25RR373 pKa = 11.84PAAAPEE379 pKa = 4.16SHH381 pKa = 5.89STRR384 pKa = 11.84PP385 pKa = 3.3
MM1 pKa = 8.12DD2 pKa = 4.81SLGALAAQFGIANLFGRR19 pKa = 11.84GFISVIITFFAAWVFTWQFIPRR41 pKa = 11.84LRR43 pKa = 11.84DD44 pKa = 3.26FAVKK48 pKa = 10.32AGWADD53 pKa = 3.35QPNARR58 pKa = 11.84RR59 pKa = 11.84LNKK62 pKa = 10.16EE63 pKa = 3.86PLPNAGGLAIFAGFITSVVLAWALRR88 pKa = 11.84PIMVEE93 pKa = 3.66LVNIQVLSILLGASIMVLVGFIDD116 pKa = 3.86DD117 pKa = 3.94QFGLSPVTRR126 pKa = 11.84LVVQVLVAVLLVVNGLKK143 pKa = 9.43MDD145 pKa = 4.5FNAIPFLPVLPEE157 pKa = 3.99AVNEE161 pKa = 4.05PLSTLLTIVWIVGLTNAVNLMDD183 pKa = 4.56GVDD186 pKa = 3.82GVVGGVGFVVSMVLLVTAAQFTDD209 pKa = 3.37RR210 pKa = 11.84AAAVILLAGLAGATLGYY227 pKa = 10.07LRR229 pKa = 11.84HH230 pKa = 6.18NFNPSRR236 pKa = 11.84IIMGDD241 pKa = 2.77AGAYY245 pKa = 10.48LIGFTLAAVSLLGTLKK261 pKa = 10.5FSAGASLIVPLIVLALPVLDD281 pKa = 3.76TTQVVIGRR289 pKa = 11.84LARR292 pKa = 11.84GIRR295 pKa = 11.84NPLGHH300 pKa = 7.52PDD302 pKa = 3.16KK303 pKa = 10.5THH305 pKa = 4.92IHH307 pKa = 5.91HH308 pKa = 6.94RR309 pKa = 11.84VLARR313 pKa = 11.84TASARR318 pKa = 11.84RR319 pKa = 11.84TAVILWMVALACGMLGMLLQGVPLLAIGLTGVLVLVCLWFVAHH362 pKa = 6.21RR363 pKa = 11.84RR364 pKa = 11.84VRR366 pKa = 11.84AHH368 pKa = 5.87SRR370 pKa = 11.84EE371 pKa = 4.12DD372 pKa = 3.25RR373 pKa = 11.84PAAAPEE379 pKa = 4.16SHH381 pKa = 5.89STRR384 pKa = 11.84PP385 pKa = 3.3
Molecular weight: 41.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1081085 |
10 |
3511 |
312.5 |
33.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.87 ± 0.059 | 0.616 ± 0.011 |
5.063 ± 0.032 | 5.596 ± 0.05 |
3.155 ± 0.024 | 8.769 ± 0.046 |
2.331 ± 0.024 | 3.687 ± 0.036 |
2.49 ± 0.035 | 11.513 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.081 ± 0.02 | 2.561 ± 0.03 |
5.646 ± 0.036 | 4.069 ± 0.029 |
7.327 ± 0.044 | 5.397 ± 0.03 |
6.159 ± 0.044 | 7.993 ± 0.035 |
1.349 ± 0.019 | 2.329 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
