
Acidaminobacter hydrogenoformans DSM 2784
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Eubacteriales incertae sedis; Eubacteriales Family XII. Incertae Sedis; Acidaminobacter; Acidaminobacter hydrogenoformans
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
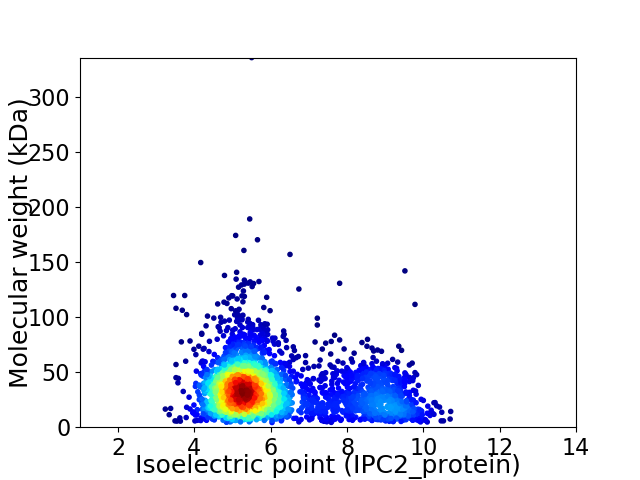
Virtual 2D-PAGE plot for 3277 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G5RQP3|A0A1G5RQP3_9FIRM Aminobenzoyl-glutamate transport protein OS=Acidaminobacter hydrogenoformans DSM 2784 OX=1120920 GN=SAMN03080599_00228 PE=4 SV=1
MM1 pKa = 7.14KK2 pKa = 10.23RR3 pKa = 11.84SSRR6 pKa = 11.84FLAISLSLMLVFQAFVIAPISWAADD31 pKa = 3.23FDD33 pKa = 3.9ITLDD37 pKa = 3.74GSQSATVNNAIFKK50 pKa = 9.91QSPSDD55 pKa = 3.46SSTGTGIFTPFVRR68 pKa = 11.84VQGNGTEE75 pKa = 4.07SGYY78 pKa = 8.73NTDD81 pKa = 3.29GTTEE85 pKa = 4.21FDD87 pKa = 3.51TKK89 pKa = 9.45TGKK92 pKa = 6.86WTHH95 pKa = 6.69AIKK98 pKa = 10.76LSDD101 pKa = 3.62IPTVDD106 pKa = 3.41VQGKK110 pKa = 8.32FYY112 pKa = 11.16RR113 pKa = 11.84EE114 pKa = 3.75LLLDD118 pKa = 4.14LNEE121 pKa = 4.63TKK123 pKa = 10.56NDD125 pKa = 3.69PYY127 pKa = 10.83IQLDD131 pKa = 3.72EE132 pKa = 4.56FEE134 pKa = 4.35IWLTKK139 pKa = 10.86DD140 pKa = 3.35KK141 pKa = 10.84DD142 pKa = 3.25ISGYY146 pKa = 10.11PFTTDD151 pKa = 2.81QAIMVFSLEE160 pKa = 4.23GEE162 pKa = 4.56SILLDD167 pKa = 3.42YY168 pKa = 10.58RR169 pKa = 11.84INSGSGWGDD178 pKa = 3.4YY179 pKa = 10.14KK180 pKa = 11.44ALIPDD185 pKa = 4.0EE186 pKa = 5.15LFAGYY191 pKa = 10.89GNDD194 pKa = 3.28YY195 pKa = 11.01YY196 pKa = 11.13FVLFSKK202 pKa = 9.19LTQADD207 pKa = 3.59SGFEE211 pKa = 3.79EE212 pKa = 4.49WGVLSSDD219 pKa = 2.61ITAAPSVDD227 pKa = 3.15IEE229 pKa = 4.23KK230 pKa = 10.63LISIDD235 pKa = 5.84DD236 pKa = 4.08GEE238 pKa = 4.52TWLDD242 pKa = 3.28ADD244 pKa = 3.96TSEE247 pKa = 4.22DD248 pKa = 3.5TVLAVYY254 pKa = 9.94PGTVHH259 pKa = 6.68FKK261 pKa = 11.02VIIEE265 pKa = 4.14NTGNVPLTNVAVTDD279 pKa = 4.39FSDD282 pKa = 3.81DD283 pKa = 3.55SEE285 pKa = 6.77AIDD288 pKa = 3.73FTGLNDD294 pKa = 3.65YY295 pKa = 10.95LGLGDD300 pKa = 5.46KK301 pKa = 10.59IEE303 pKa = 4.48SDD305 pKa = 4.38PIEE308 pKa = 4.18VSTILRR314 pKa = 11.84SHH316 pKa = 6.99ANTATINAYY325 pKa = 9.52YY326 pKa = 10.32GKK328 pKa = 10.49EE329 pKa = 3.9KK330 pKa = 10.33VTDD333 pKa = 3.65SDD335 pKa = 3.73SANYY339 pKa = 7.52YY340 pKa = 7.74TLPDD344 pKa = 3.79PKK346 pKa = 10.36IAINKK351 pKa = 5.86TTNGDD356 pKa = 3.63DD357 pKa = 3.94DD358 pKa = 5.99LDD360 pKa = 5.61IIVGSDD366 pKa = 3.34VTWEE370 pKa = 4.17YY371 pKa = 11.23EE372 pKa = 3.91VTNIGNVPLSGISVNDD388 pKa = 3.08SDD390 pKa = 5.51EE391 pKa = 4.45NVTPAYY397 pKa = 10.76VSGDD401 pKa = 3.55TNEE404 pKa = 5.7DD405 pKa = 3.39DD406 pKa = 4.55MLDD409 pKa = 3.66LTEE412 pKa = 3.88TWIFKK417 pKa = 10.75ASGTAIQGSYY427 pKa = 11.08SNTGYY432 pKa = 11.03VSGTFEE438 pKa = 4.44EE439 pKa = 4.93STATADD445 pKa = 4.42DD446 pKa = 3.97DD447 pKa = 4.9SNYY450 pKa = 10.4FGYY453 pKa = 10.32IPDD456 pKa = 4.09APAIQITKK464 pKa = 6.43TTNGDD469 pKa = 3.39DD470 pKa = 3.65GLEE473 pKa = 3.94IEE475 pKa = 4.89VGSEE479 pKa = 3.75ITWSYY484 pKa = 11.21EE485 pKa = 3.49VTNKK489 pKa = 10.47GNVALGNITVTDD501 pKa = 3.84SDD503 pKa = 3.76EE504 pKa = 5.35SITPVYY510 pKa = 10.87VSGDD514 pKa = 3.43ANTDD518 pKa = 3.14GNLDD522 pKa = 3.62LTEE525 pKa = 3.92TWIYY529 pKa = 10.2EE530 pKa = 4.16ATGTAIEE537 pKa = 4.49GNYY540 pKa = 10.44SNTGYY545 pKa = 11.05VSGTFEE551 pKa = 3.76EE552 pKa = 4.4ATYY555 pKa = 10.95RR556 pKa = 11.84DD557 pKa = 4.94DD558 pKa = 5.95DD559 pKa = 4.63DD560 pKa = 4.93SSYY563 pKa = 11.23FGYY566 pKa = 10.41IPDD569 pKa = 4.84DD570 pKa = 3.61PAIQIIKK577 pKa = 6.19TTNGNDD583 pKa = 3.45GLEE586 pKa = 4.05IEE588 pKa = 4.77VGSEE592 pKa = 3.44ITWRR596 pKa = 11.84YY597 pKa = 9.68EE598 pKa = 3.34ITNKK602 pKa = 10.42GNVALGNITVTDD614 pKa = 3.84SDD616 pKa = 3.76EE617 pKa = 5.35SITPVYY623 pKa = 10.87VSGDD627 pKa = 3.38ANTDD631 pKa = 2.9GKK633 pKa = 11.33LDD635 pKa = 4.12LKK637 pKa = 8.88EE638 pKa = 3.88TWIYY642 pKa = 10.39EE643 pKa = 4.17ATGTAIEE650 pKa = 4.56GNYY653 pKa = 10.55SNIGYY658 pKa = 10.07VSGSFGEE665 pKa = 4.6GTVNDD670 pKa = 4.49NDD672 pKa = 3.4DD673 pKa = 3.72SYY675 pKa = 10.37YY676 pKa = 10.36TGYY679 pKa = 10.55VPQEE683 pKa = 4.02PKK685 pKa = 10.26TPEE688 pKa = 3.14ISINKK693 pKa = 5.92TTNGSDD699 pKa = 3.3GSRR702 pKa = 11.84IRR704 pKa = 11.84VGTAITWEE712 pKa = 4.34YY713 pKa = 10.98KK714 pKa = 10.25VEE716 pKa = 4.19TVNGFDD722 pKa = 3.98IYY724 pKa = 11.4DD725 pKa = 3.61VDD727 pKa = 4.16VTDD730 pKa = 5.33SDD732 pKa = 4.05TDD734 pKa = 3.96LDD736 pKa = 3.9PQYY739 pKa = 11.79VSGDD743 pKa = 3.65DD744 pKa = 3.65GDD746 pKa = 5.1GVLEE750 pKa = 4.88PGEE753 pKa = 4.09TWIYY757 pKa = 9.95QATGTAVSGLYY768 pKa = 10.45EE769 pKa = 4.38NIGYY773 pKa = 7.51VTGYY777 pKa = 10.72AEE779 pKa = 4.26RR780 pKa = 11.84EE781 pKa = 4.3VPVSDD786 pKa = 4.81EE787 pKa = 4.27DD788 pKa = 3.86TSSYY792 pKa = 10.19TGYY795 pKa = 8.75TVTTTTTTRR804 pKa = 11.84RR805 pKa = 11.84TPDD808 pKa = 3.95PDD810 pKa = 4.09PGNIEE815 pKa = 3.56IFKK818 pKa = 10.9FLDD821 pKa = 3.28SDD823 pKa = 4.21GNGSFDD829 pKa = 4.31EE830 pKa = 4.35EE831 pKa = 4.55NEE833 pKa = 4.18FPFEE837 pKa = 4.3NIRR840 pKa = 11.84FQLYY844 pKa = 10.8DD845 pKa = 3.64EE846 pKa = 5.02DD847 pKa = 4.34KK848 pKa = 11.46GLIEE852 pKa = 4.15TASTDD857 pKa = 3.71DD858 pKa = 3.48EE859 pKa = 4.83GMLTFRR865 pKa = 11.84NLDD868 pKa = 3.15AGTYY872 pKa = 8.58YY873 pKa = 10.14IKK875 pKa = 10.4EE876 pKa = 3.88ARR878 pKa = 11.84NDD880 pKa = 3.5YY881 pKa = 10.94SITTGGFDD889 pKa = 3.26EE890 pKa = 4.62NGFYY894 pKa = 10.62EE895 pKa = 4.25VDD897 pKa = 3.23VDD899 pKa = 3.64EE900 pKa = 6.0DD901 pKa = 3.76EE902 pKa = 4.42TVEE905 pKa = 4.28IEE907 pKa = 3.93VGNYY911 pKa = 9.72RR912 pKa = 11.84EE913 pKa = 4.34VIPPQEE919 pKa = 4.67PPLGPPPEE927 pKa = 4.04VEE929 pKa = 4.44EE930 pKa = 3.99ILEE933 pKa = 4.27EE934 pKa = 4.27KK935 pKa = 10.43VPQGPPLPQTGEE947 pKa = 3.8IPPYY951 pKa = 8.51FAYY954 pKa = 10.22GIGSLLILAGAFMRR968 pKa = 11.84RR969 pKa = 11.84KK970 pKa = 9.42FF971 pKa = 3.46
MM1 pKa = 7.14KK2 pKa = 10.23RR3 pKa = 11.84SSRR6 pKa = 11.84FLAISLSLMLVFQAFVIAPISWAADD31 pKa = 3.23FDD33 pKa = 3.9ITLDD37 pKa = 3.74GSQSATVNNAIFKK50 pKa = 9.91QSPSDD55 pKa = 3.46SSTGTGIFTPFVRR68 pKa = 11.84VQGNGTEE75 pKa = 4.07SGYY78 pKa = 8.73NTDD81 pKa = 3.29GTTEE85 pKa = 4.21FDD87 pKa = 3.51TKK89 pKa = 9.45TGKK92 pKa = 6.86WTHH95 pKa = 6.69AIKK98 pKa = 10.76LSDD101 pKa = 3.62IPTVDD106 pKa = 3.41VQGKK110 pKa = 8.32FYY112 pKa = 11.16RR113 pKa = 11.84EE114 pKa = 3.75LLLDD118 pKa = 4.14LNEE121 pKa = 4.63TKK123 pKa = 10.56NDD125 pKa = 3.69PYY127 pKa = 10.83IQLDD131 pKa = 3.72EE132 pKa = 4.56FEE134 pKa = 4.35IWLTKK139 pKa = 10.86DD140 pKa = 3.35KK141 pKa = 10.84DD142 pKa = 3.25ISGYY146 pKa = 10.11PFTTDD151 pKa = 2.81QAIMVFSLEE160 pKa = 4.23GEE162 pKa = 4.56SILLDD167 pKa = 3.42YY168 pKa = 10.58RR169 pKa = 11.84INSGSGWGDD178 pKa = 3.4YY179 pKa = 10.14KK180 pKa = 11.44ALIPDD185 pKa = 4.0EE186 pKa = 5.15LFAGYY191 pKa = 10.89GNDD194 pKa = 3.28YY195 pKa = 11.01YY196 pKa = 11.13FVLFSKK202 pKa = 9.19LTQADD207 pKa = 3.59SGFEE211 pKa = 3.79EE212 pKa = 4.49WGVLSSDD219 pKa = 2.61ITAAPSVDD227 pKa = 3.15IEE229 pKa = 4.23KK230 pKa = 10.63LISIDD235 pKa = 5.84DD236 pKa = 4.08GEE238 pKa = 4.52TWLDD242 pKa = 3.28ADD244 pKa = 3.96TSEE247 pKa = 4.22DD248 pKa = 3.5TVLAVYY254 pKa = 9.94PGTVHH259 pKa = 6.68FKK261 pKa = 11.02VIIEE265 pKa = 4.14NTGNVPLTNVAVTDD279 pKa = 4.39FSDD282 pKa = 3.81DD283 pKa = 3.55SEE285 pKa = 6.77AIDD288 pKa = 3.73FTGLNDD294 pKa = 3.65YY295 pKa = 10.95LGLGDD300 pKa = 5.46KK301 pKa = 10.59IEE303 pKa = 4.48SDD305 pKa = 4.38PIEE308 pKa = 4.18VSTILRR314 pKa = 11.84SHH316 pKa = 6.99ANTATINAYY325 pKa = 9.52YY326 pKa = 10.32GKK328 pKa = 10.49EE329 pKa = 3.9KK330 pKa = 10.33VTDD333 pKa = 3.65SDD335 pKa = 3.73SANYY339 pKa = 7.52YY340 pKa = 7.74TLPDD344 pKa = 3.79PKK346 pKa = 10.36IAINKK351 pKa = 5.86TTNGDD356 pKa = 3.63DD357 pKa = 3.94DD358 pKa = 5.99LDD360 pKa = 5.61IIVGSDD366 pKa = 3.34VTWEE370 pKa = 4.17YY371 pKa = 11.23EE372 pKa = 3.91VTNIGNVPLSGISVNDD388 pKa = 3.08SDD390 pKa = 5.51EE391 pKa = 4.45NVTPAYY397 pKa = 10.76VSGDD401 pKa = 3.55TNEE404 pKa = 5.7DD405 pKa = 3.39DD406 pKa = 4.55MLDD409 pKa = 3.66LTEE412 pKa = 3.88TWIFKK417 pKa = 10.75ASGTAIQGSYY427 pKa = 11.08SNTGYY432 pKa = 11.03VSGTFEE438 pKa = 4.44EE439 pKa = 4.93STATADD445 pKa = 4.42DD446 pKa = 3.97DD447 pKa = 4.9SNYY450 pKa = 10.4FGYY453 pKa = 10.32IPDD456 pKa = 4.09APAIQITKK464 pKa = 6.43TTNGDD469 pKa = 3.39DD470 pKa = 3.65GLEE473 pKa = 3.94IEE475 pKa = 4.89VGSEE479 pKa = 3.75ITWSYY484 pKa = 11.21EE485 pKa = 3.49VTNKK489 pKa = 10.47GNVALGNITVTDD501 pKa = 3.84SDD503 pKa = 3.76EE504 pKa = 5.35SITPVYY510 pKa = 10.87VSGDD514 pKa = 3.43ANTDD518 pKa = 3.14GNLDD522 pKa = 3.62LTEE525 pKa = 3.92TWIYY529 pKa = 10.2EE530 pKa = 4.16ATGTAIEE537 pKa = 4.49GNYY540 pKa = 10.44SNTGYY545 pKa = 11.05VSGTFEE551 pKa = 3.76EE552 pKa = 4.4ATYY555 pKa = 10.95RR556 pKa = 11.84DD557 pKa = 4.94DD558 pKa = 5.95DD559 pKa = 4.63DD560 pKa = 4.93SSYY563 pKa = 11.23FGYY566 pKa = 10.41IPDD569 pKa = 4.84DD570 pKa = 3.61PAIQIIKK577 pKa = 6.19TTNGNDD583 pKa = 3.45GLEE586 pKa = 4.05IEE588 pKa = 4.77VGSEE592 pKa = 3.44ITWRR596 pKa = 11.84YY597 pKa = 9.68EE598 pKa = 3.34ITNKK602 pKa = 10.42GNVALGNITVTDD614 pKa = 3.84SDD616 pKa = 3.76EE617 pKa = 5.35SITPVYY623 pKa = 10.87VSGDD627 pKa = 3.38ANTDD631 pKa = 2.9GKK633 pKa = 11.33LDD635 pKa = 4.12LKK637 pKa = 8.88EE638 pKa = 3.88TWIYY642 pKa = 10.39EE643 pKa = 4.17ATGTAIEE650 pKa = 4.56GNYY653 pKa = 10.55SNIGYY658 pKa = 10.07VSGSFGEE665 pKa = 4.6GTVNDD670 pKa = 4.49NDD672 pKa = 3.4DD673 pKa = 3.72SYY675 pKa = 10.37YY676 pKa = 10.36TGYY679 pKa = 10.55VPQEE683 pKa = 4.02PKK685 pKa = 10.26TPEE688 pKa = 3.14ISINKK693 pKa = 5.92TTNGSDD699 pKa = 3.3GSRR702 pKa = 11.84IRR704 pKa = 11.84VGTAITWEE712 pKa = 4.34YY713 pKa = 10.98KK714 pKa = 10.25VEE716 pKa = 4.19TVNGFDD722 pKa = 3.98IYY724 pKa = 11.4DD725 pKa = 3.61VDD727 pKa = 4.16VTDD730 pKa = 5.33SDD732 pKa = 4.05TDD734 pKa = 3.96LDD736 pKa = 3.9PQYY739 pKa = 11.79VSGDD743 pKa = 3.65DD744 pKa = 3.65GDD746 pKa = 5.1GVLEE750 pKa = 4.88PGEE753 pKa = 4.09TWIYY757 pKa = 9.95QATGTAVSGLYY768 pKa = 10.45EE769 pKa = 4.38NIGYY773 pKa = 7.51VTGYY777 pKa = 10.72AEE779 pKa = 4.26RR780 pKa = 11.84EE781 pKa = 4.3VPVSDD786 pKa = 4.81EE787 pKa = 4.27DD788 pKa = 3.86TSSYY792 pKa = 10.19TGYY795 pKa = 8.75TVTTTTTTRR804 pKa = 11.84RR805 pKa = 11.84TPDD808 pKa = 3.95PDD810 pKa = 4.09PGNIEE815 pKa = 3.56IFKK818 pKa = 10.9FLDD821 pKa = 3.28SDD823 pKa = 4.21GNGSFDD829 pKa = 4.31EE830 pKa = 4.35EE831 pKa = 4.55NEE833 pKa = 4.18FPFEE837 pKa = 4.3NIRR840 pKa = 11.84FQLYY844 pKa = 10.8DD845 pKa = 3.64EE846 pKa = 5.02DD847 pKa = 4.34KK848 pKa = 11.46GLIEE852 pKa = 4.15TASTDD857 pKa = 3.71DD858 pKa = 3.48EE859 pKa = 4.83GMLTFRR865 pKa = 11.84NLDD868 pKa = 3.15AGTYY872 pKa = 8.58YY873 pKa = 10.14IKK875 pKa = 10.4EE876 pKa = 3.88ARR878 pKa = 11.84NDD880 pKa = 3.5YY881 pKa = 10.94SITTGGFDD889 pKa = 3.26EE890 pKa = 4.62NGFYY894 pKa = 10.62EE895 pKa = 4.25VDD897 pKa = 3.23VDD899 pKa = 3.64EE900 pKa = 6.0DD901 pKa = 3.76EE902 pKa = 4.42TVEE905 pKa = 4.28IEE907 pKa = 3.93VGNYY911 pKa = 9.72RR912 pKa = 11.84EE913 pKa = 4.34VIPPQEE919 pKa = 4.67PPLGPPPEE927 pKa = 4.04VEE929 pKa = 4.44EE930 pKa = 3.99ILEE933 pKa = 4.27EE934 pKa = 4.27KK935 pKa = 10.43VPQGPPLPQTGEE947 pKa = 3.8IPPYY951 pKa = 8.51FAYY954 pKa = 10.22GIGSLLILAGAFMRR968 pKa = 11.84RR969 pKa = 11.84KK970 pKa = 9.42FF971 pKa = 3.46
Molecular weight: 106.25 kDa
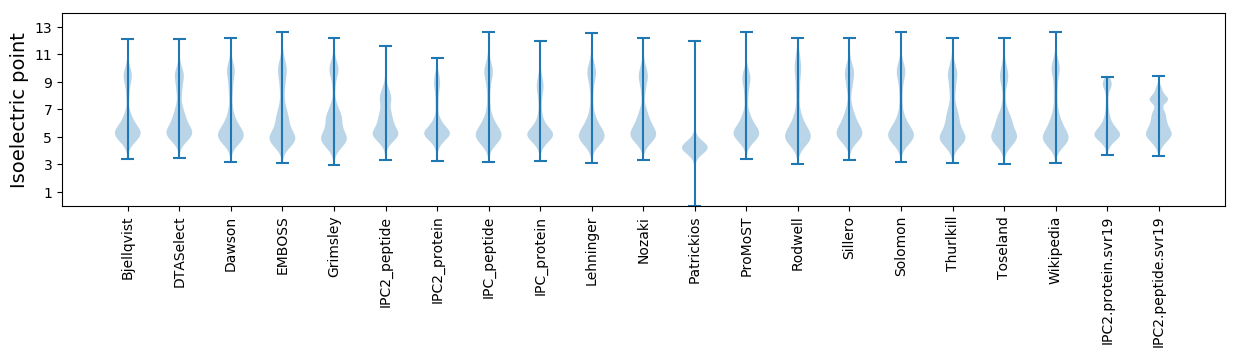
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G5S6R0|A0A1G5S6R0_9FIRM Uncharacterized protein OS=Acidaminobacter hydrogenoformans DSM 2784 OX=1120920 GN=SAMN03080599_03247 PE=4 SV=1
AA1 pKa = 7.98CITAFRR7 pKa = 11.84NWSKK11 pKa = 11.06EE12 pKa = 3.21ILNAFKK18 pKa = 10.81YY19 pKa = 10.34GYY21 pKa = 9.31TNGCTEE27 pKa = 4.25GFNNKK32 pKa = 8.96IKK34 pKa = 9.61VLKK37 pKa = 9.9RR38 pKa = 11.84ISYY41 pKa = 8.63GVRR44 pKa = 11.84NFMRR48 pKa = 11.84FRR50 pKa = 11.84NRR52 pKa = 11.84ILHH55 pKa = 4.88MCRR58 pKa = 3.49
AA1 pKa = 7.98CITAFRR7 pKa = 11.84NWSKK11 pKa = 11.06EE12 pKa = 3.21ILNAFKK18 pKa = 10.81YY19 pKa = 10.34GYY21 pKa = 9.31TNGCTEE27 pKa = 4.25GFNNKK32 pKa = 8.96IKK34 pKa = 9.61VLKK37 pKa = 9.9RR38 pKa = 11.84ISYY41 pKa = 8.63GVRR44 pKa = 11.84NFMRR48 pKa = 11.84FRR50 pKa = 11.84NRR52 pKa = 11.84ILHH55 pKa = 4.88MCRR58 pKa = 3.49
Molecular weight: 6.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1050301 |
40 |
2971 |
320.5 |
35.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.57 ± 0.048 | 0.945 ± 0.018 |
5.287 ± 0.037 | 7.245 ± 0.043 |
4.18 ± 0.029 | 7.59 ± 0.043 |
1.888 ± 0.019 | 7.172 ± 0.042 |
5.85 ± 0.036 | 10.127 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.051 ± 0.021 | 3.668 ± 0.029 |
3.831 ± 0.026 | 3.014 ± 0.022 |
4.719 ± 0.036 | 6.003 ± 0.031 |
5.297 ± 0.031 | 7.384 ± 0.034 |
0.844 ± 0.014 | 3.335 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
