
Bison bison bison
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Mammalia; Theria; Eutheria; Boreoeutheria; Laurasiatheria; Artiodactyla;
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
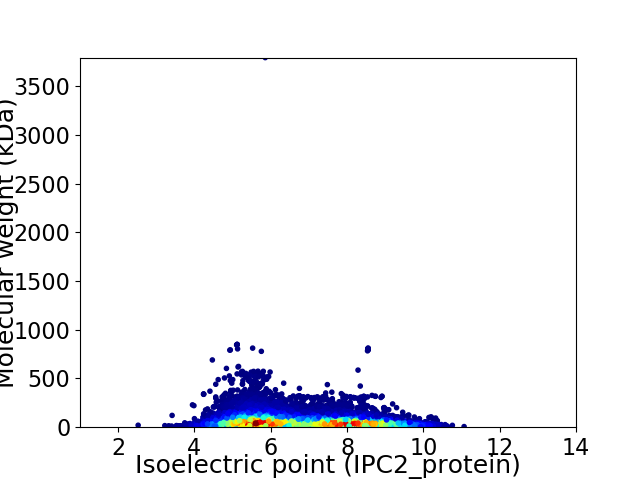
Virtual 2D-PAGE plot for 29577 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6P3GQ86|A0A6P3GQ86_BISBI melanoma-associated antigen 10-like OS=Bison bison bison OX=43346 GN=LOC104983845 PE=4 SV=1
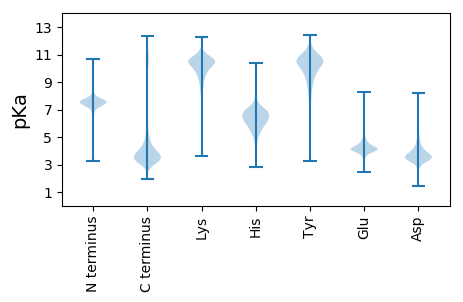
GG1 pKa = 8.25DD2 pKa = 3.65IQQLLLVSDD11 pKa = 4.08HH12 pKa = 6.57RR13 pKa = 11.84AAYY16 pKa = 9.28DD17 pKa = 3.56YY18 pKa = 10.97CEE20 pKa = 5.09HH21 pKa = 6.88YY22 pKa = 11.21SPDD25 pKa = 3.94CDD27 pKa = 3.16TAVPDD32 pKa = 4.49KK33 pKa = 10.74PQSQDD38 pKa = 3.47PNPDD42 pKa = 3.48EE43 pKa = 5.17YY44 pKa = 11.27EE45 pKa = 3.96LTQPPTEE52 pKa = 4.55APSAPSTTEE61 pKa = 3.92GPGKK65 pKa = 10.61EE66 pKa = 4.09EE67 pKa = 4.34DD68 pKa = 4.2PGIGDD73 pKa = 3.71YY74 pKa = 10.99DD75 pKa = 3.92YY76 pKa = 11.74VPSEE80 pKa = 5.03DD81 pKa = 5.21YY82 pKa = 8.57YY83 pKa = 11.2TPPPYY88 pKa = 10.55EE89 pKa = 4.01DD90 pKa = 3.07FGYY93 pKa = 11.22GEE95 pKa = 5.12GIEE98 pKa = 4.87NPDD101 pKa = 4.02EE102 pKa = 5.22NPDD105 pKa = 3.58QLPDD109 pKa = 3.14QGARR113 pKa = 11.84AEE115 pKa = 4.54VPTSTVLTSNGSNPAPPPEE134 pKa = 4.28EE135 pKa = 5.77GIDD138 pKa = 3.7DD139 pKa = 4.1LEE141 pKa = 4.46EE142 pKa = 4.14EE143 pKa = 4.53FTEE146 pKa = 4.24EE147 pKa = 4.01TIKK150 pKa = 11.11NLDD153 pKa = 3.18EE154 pKa = 4.85SYY156 pKa = 11.05YY157 pKa = 11.07DD158 pKa = 3.68PYY160 pKa = 11.27YY161 pKa = 11.04DD162 pKa = 3.66PTISPSEE169 pKa = 4.13IGPGMPANQDD179 pKa = 3.51TIYY182 pKa = 10.65EE183 pKa = 4.36GMGGPRR189 pKa = 11.84GEE191 pKa = 4.5KK192 pKa = 9.01GQKK195 pKa = 9.93GEE197 pKa = 4.14PAIIEE202 pKa = 4.22PGMLLEE208 pKa = 5.04GPPGPEE214 pKa = 4.27GPAGLPGPPGTMGPTGQVGDD234 pKa = 4.16PGEE237 pKa = 4.51RR238 pKa = 11.84GPPGRR243 pKa = 11.84PGLPGADD250 pKa = 3.87GLPGPPGTMLMLPFRR265 pKa = 11.84FGGGGDD271 pKa = 3.7AGSKK275 pKa = 10.78GPMVSAQEE283 pKa = 4.2SQAQAILQQARR294 pKa = 11.84VSGLGHH300 pKa = 6.96GGRR303 pKa = 11.84LGAQLARR310 pKa = 11.84LLTWGPRR317 pKa = 11.84FPDD320 pKa = 2.94MDD322 pKa = 3.82MAAEE326 pKa = 4.18PLWLLLTPSSSTIYY340 pKa = 10.15CLPPGSGMDD349 pKa = 3.47RR350 pKa = 11.84PHH352 pKa = 7.14VLLGGPLL359 pKa = 3.29
GG1 pKa = 8.25DD2 pKa = 3.65IQQLLLVSDD11 pKa = 4.08HH12 pKa = 6.57RR13 pKa = 11.84AAYY16 pKa = 9.28DD17 pKa = 3.56YY18 pKa = 10.97CEE20 pKa = 5.09HH21 pKa = 6.88YY22 pKa = 11.21SPDD25 pKa = 3.94CDD27 pKa = 3.16TAVPDD32 pKa = 4.49KK33 pKa = 10.74PQSQDD38 pKa = 3.47PNPDD42 pKa = 3.48EE43 pKa = 5.17YY44 pKa = 11.27EE45 pKa = 3.96LTQPPTEE52 pKa = 4.55APSAPSTTEE61 pKa = 3.92GPGKK65 pKa = 10.61EE66 pKa = 4.09EE67 pKa = 4.34DD68 pKa = 4.2PGIGDD73 pKa = 3.71YY74 pKa = 10.99DD75 pKa = 3.92YY76 pKa = 11.74VPSEE80 pKa = 5.03DD81 pKa = 5.21YY82 pKa = 8.57YY83 pKa = 11.2TPPPYY88 pKa = 10.55EE89 pKa = 4.01DD90 pKa = 3.07FGYY93 pKa = 11.22GEE95 pKa = 5.12GIEE98 pKa = 4.87NPDD101 pKa = 4.02EE102 pKa = 5.22NPDD105 pKa = 3.58QLPDD109 pKa = 3.14QGARR113 pKa = 11.84AEE115 pKa = 4.54VPTSTVLTSNGSNPAPPPEE134 pKa = 4.28EE135 pKa = 5.77GIDD138 pKa = 3.7DD139 pKa = 4.1LEE141 pKa = 4.46EE142 pKa = 4.14EE143 pKa = 4.53FTEE146 pKa = 4.24EE147 pKa = 4.01TIKK150 pKa = 11.11NLDD153 pKa = 3.18EE154 pKa = 4.85SYY156 pKa = 11.05YY157 pKa = 11.07DD158 pKa = 3.68PYY160 pKa = 11.27YY161 pKa = 11.04DD162 pKa = 3.66PTISPSEE169 pKa = 4.13IGPGMPANQDD179 pKa = 3.51TIYY182 pKa = 10.65EE183 pKa = 4.36GMGGPRR189 pKa = 11.84GEE191 pKa = 4.5KK192 pKa = 9.01GQKK195 pKa = 9.93GEE197 pKa = 4.14PAIIEE202 pKa = 4.22PGMLLEE208 pKa = 5.04GPPGPEE214 pKa = 4.27GPAGLPGPPGTMGPTGQVGDD234 pKa = 4.16PGEE237 pKa = 4.51RR238 pKa = 11.84GPPGRR243 pKa = 11.84PGLPGADD250 pKa = 3.87GLPGPPGTMLMLPFRR265 pKa = 11.84FGGGGDD271 pKa = 3.7AGSKK275 pKa = 10.78GPMVSAQEE283 pKa = 4.2SQAQAILQQARR294 pKa = 11.84VSGLGHH300 pKa = 6.96GGRR303 pKa = 11.84LGAQLARR310 pKa = 11.84LLTWGPRR317 pKa = 11.84FPDD320 pKa = 2.94MDD322 pKa = 3.82MAAEE326 pKa = 4.18PLWLLLTPSSSTIYY340 pKa = 10.15CLPPGSGMDD349 pKa = 3.47RR350 pKa = 11.84PHH352 pKa = 7.14VLLGGPLL359 pKa = 3.29
Molecular weight: 37.71 kDa
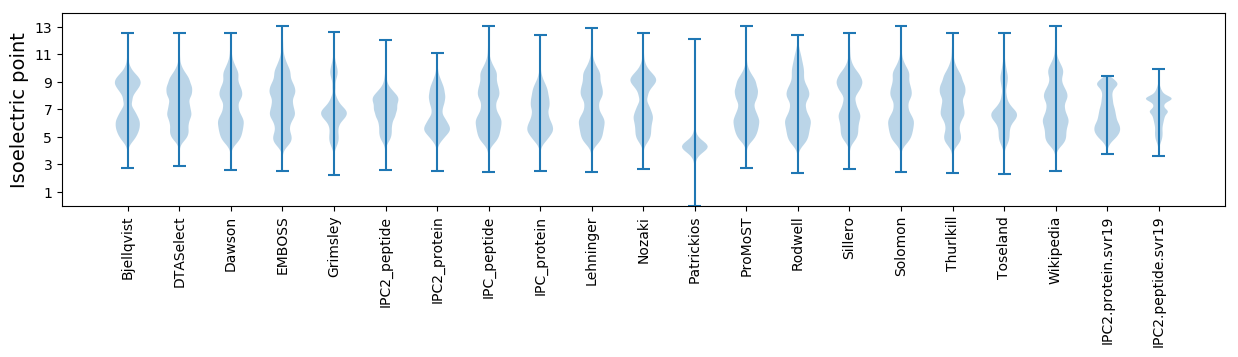
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6P3HRY6|A0A6P3HRY6_BISBI FERM and PDZ domain-containing protein 1 OS=Bison bison bison OX=43346 GN=FRMPD1 PE=4 SV=1
MM1 pKa = 7.57SSHH4 pKa = 5.15KK5 pKa = 8.91TFRR8 pKa = 11.84IKK10 pKa = 10.64RR11 pKa = 11.84FLAKK15 pKa = 9.71KK16 pKa = 9.58QKK18 pKa = 8.69QNRR21 pKa = 11.84PIPQWIRR28 pKa = 11.84MKK30 pKa = 9.89TGNKK34 pKa = 8.61IRR36 pKa = 11.84YY37 pKa = 7.09NSKK40 pKa = 8.3RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 3.95WRR45 pKa = 11.84RR46 pKa = 11.84TKK48 pKa = 10.83LGLL51 pKa = 3.67
MM1 pKa = 7.57SSHH4 pKa = 5.15KK5 pKa = 8.91TFRR8 pKa = 11.84IKK10 pKa = 10.64RR11 pKa = 11.84FLAKK15 pKa = 9.71KK16 pKa = 9.58QKK18 pKa = 8.69QNRR21 pKa = 11.84PIPQWIRR28 pKa = 11.84MKK30 pKa = 9.89TGNKK34 pKa = 8.61IRR36 pKa = 11.84YY37 pKa = 7.09NSKK40 pKa = 8.3RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 3.95WRR45 pKa = 11.84RR46 pKa = 11.84TKK48 pKa = 10.83LGLL51 pKa = 3.67
Molecular weight: 6.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
17611492 |
31 |
34175 |
595.4 |
66.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.815 ± 0.013 | 2.182 ± 0.011 |
4.839 ± 0.011 | 7.17 ± 0.02 |
3.627 ± 0.009 | 6.314 ± 0.019 |
2.578 ± 0.007 | 4.394 ± 0.011 |
5.872 ± 0.02 | 9.96 ± 0.02 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.151 ± 0.007 | 3.623 ± 0.009 |
6.23 ± 0.02 | 4.836 ± 0.013 |
5.634 ± 0.012 | 8.546 ± 0.017 |
5.343 ± 0.012 | 6.032 ± 0.011 |
1.187 ± 0.006 | 2.63 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
