
Oceanispirochaeta crateris
Taxonomy: cellular organisms; Bacteria; Spirochaetes; Spirochaetia; Spirochaetales; Spirochaetaceae; Oceanispirochaeta
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
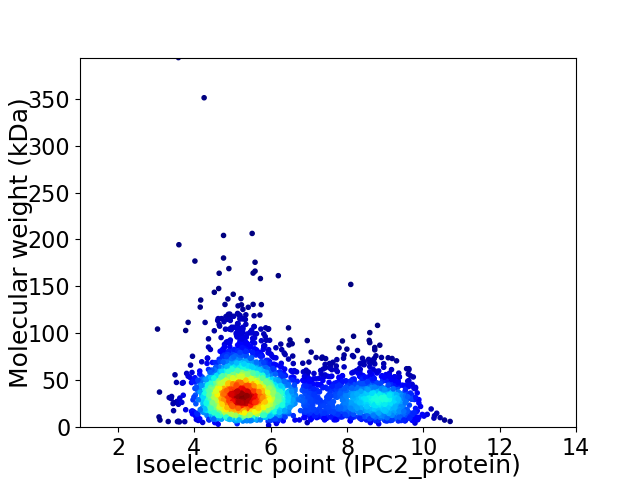
Virtual 2D-PAGE plot for 3504 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C1QMT4|A0A5C1QMT4_9SPIO FadR family transcriptional regulator OS=Oceanispirochaeta crateris OX=2518645 GN=EXM22_09825 PE=4 SV=1
MM1 pKa = 7.44NLFKK5 pKa = 10.82SAVIAAVLGVSITVIGCQTNGKK27 pKa = 8.91EE28 pKa = 4.15LTQEE32 pKa = 3.92SPKK35 pKa = 10.78VKK37 pKa = 10.22FVVTDD42 pKa = 3.51AMQTKK47 pKa = 10.17FYY49 pKa = 8.24DD50 pKa = 3.28THH52 pKa = 8.11GNIIEE57 pKa = 4.28EE58 pKa = 4.6PEE60 pKa = 4.69PGDD63 pKa = 3.56PLYY66 pKa = 11.2GQDD69 pKa = 3.52AQYY72 pKa = 11.28EE73 pKa = 4.92GIRR76 pKa = 11.84PLYY79 pKa = 8.99TDD81 pKa = 4.04NGDD84 pKa = 3.66GTVTDD89 pKa = 5.08DD90 pKa = 3.65NTGLMWQQTPPKK102 pKa = 10.77DD103 pKa = 3.13KK104 pKa = 8.68MTYY107 pKa = 9.98DD108 pKa = 3.16EE109 pKa = 4.23AMEE112 pKa = 4.01YY113 pKa = 10.57VEE115 pKa = 4.85KK116 pKa = 11.22LEE118 pKa = 4.83LGGYY122 pKa = 7.02TDD124 pKa = 3.46WRR126 pKa = 11.84LPTIKK131 pKa = 10.34EE132 pKa = 4.27SFSLANLDD140 pKa = 3.48GQLNVGDD147 pKa = 4.41TNQSVPYY154 pKa = 9.66IDD156 pKa = 4.77SDD158 pKa = 4.18YY159 pKa = 11.43FDD161 pKa = 4.35FFYY164 pKa = 11.19DD165 pKa = 3.01EE166 pKa = 4.34GRR168 pKa = 11.84PYY170 pKa = 9.98TGSYY174 pKa = 5.6WTSSVTRR181 pKa = 11.84IPAEE185 pKa = 3.8NEE187 pKa = 3.72YY188 pKa = 11.34EE189 pKa = 4.16EE190 pKa = 4.35MEE192 pKa = 4.36KK193 pKa = 10.83NYY195 pKa = 10.32GFNWADD201 pKa = 3.35GHH203 pKa = 6.23LKK205 pKa = 10.58SYY207 pKa = 11.16GDD209 pKa = 4.23GYY211 pKa = 11.34SLDD214 pKa = 3.84GSTSGSSIPAGVRR227 pKa = 11.84AVRR230 pKa = 11.84GEE232 pKa = 3.9EE233 pKa = 3.9GVYY236 pKa = 10.46GVNDD240 pKa = 3.48FTDD243 pKa = 3.47NRR245 pKa = 11.84DD246 pKa = 3.29GTITDD251 pKa = 3.46QATGLMWSQKK261 pKa = 10.86DD262 pKa = 3.48SGAVNDD268 pKa = 4.75DD269 pKa = 3.34GTLRR273 pKa = 11.84SEE275 pKa = 4.16DD276 pKa = 4.1DD277 pKa = 3.6EE278 pKa = 4.89NFGLGRR284 pKa = 11.84TWVDD288 pKa = 3.08TLVWVQNMNAAQYY301 pKa = 10.73LGYY304 pKa = 10.16SDD306 pKa = 4.31WRR308 pKa = 11.84LPDD311 pKa = 3.68AKK313 pKa = 9.69EE314 pKa = 3.83LQSIVQYY321 pKa = 10.82GISEE325 pKa = 4.31LPATDD330 pKa = 3.8SDD332 pKa = 4.44YY333 pKa = 11.58FDD335 pKa = 5.05LSRR338 pKa = 11.84TDD340 pKa = 3.08SYY342 pKa = 10.88MWTSTTCGDD351 pKa = 4.09FPDD354 pKa = 3.84TALYY358 pKa = 9.95FAFGKK363 pKa = 10.6AYY365 pKa = 10.12GINLMTGGVPAGQGPEE381 pKa = 4.19GGEE384 pKa = 3.97QMGPPPEE391 pKa = 4.74DD392 pKa = 3.33VNDD395 pKa = 3.7PMEE398 pKa = 4.7NEE400 pKa = 4.07VEE402 pKa = 4.38SFVTVDD408 pKa = 2.97ATVADD413 pKa = 4.82FVDD416 pKa = 3.73THH418 pKa = 6.92GPGAMRR424 pKa = 11.84NDD426 pKa = 3.76YY427 pKa = 11.23KK428 pKa = 11.2DD429 pKa = 3.42VTGTATQAPALSRR442 pKa = 11.84EE443 pKa = 4.39LWSKK447 pKa = 11.12LYY449 pKa = 10.94GEE451 pKa = 5.07EE452 pKa = 4.12YY453 pKa = 9.94PYY455 pKa = 11.44DD456 pKa = 4.34FDD458 pKa = 5.98PSDD461 pKa = 3.51EE462 pKa = 4.25TTLFDD467 pKa = 5.62LSASEE472 pKa = 4.16NAADD476 pKa = 4.82YY477 pKa = 10.82IVLYY481 pKa = 10.7NYY483 pKa = 10.99ALLVRR488 pKa = 11.84DD489 pKa = 3.99ADD491 pKa = 3.7
MM1 pKa = 7.44NLFKK5 pKa = 10.82SAVIAAVLGVSITVIGCQTNGKK27 pKa = 8.91EE28 pKa = 4.15LTQEE32 pKa = 3.92SPKK35 pKa = 10.78VKK37 pKa = 10.22FVVTDD42 pKa = 3.51AMQTKK47 pKa = 10.17FYY49 pKa = 8.24DD50 pKa = 3.28THH52 pKa = 8.11GNIIEE57 pKa = 4.28EE58 pKa = 4.6PEE60 pKa = 4.69PGDD63 pKa = 3.56PLYY66 pKa = 11.2GQDD69 pKa = 3.52AQYY72 pKa = 11.28EE73 pKa = 4.92GIRR76 pKa = 11.84PLYY79 pKa = 8.99TDD81 pKa = 4.04NGDD84 pKa = 3.66GTVTDD89 pKa = 5.08DD90 pKa = 3.65NTGLMWQQTPPKK102 pKa = 10.77DD103 pKa = 3.13KK104 pKa = 8.68MTYY107 pKa = 9.98DD108 pKa = 3.16EE109 pKa = 4.23AMEE112 pKa = 4.01YY113 pKa = 10.57VEE115 pKa = 4.85KK116 pKa = 11.22LEE118 pKa = 4.83LGGYY122 pKa = 7.02TDD124 pKa = 3.46WRR126 pKa = 11.84LPTIKK131 pKa = 10.34EE132 pKa = 4.27SFSLANLDD140 pKa = 3.48GQLNVGDD147 pKa = 4.41TNQSVPYY154 pKa = 9.66IDD156 pKa = 4.77SDD158 pKa = 4.18YY159 pKa = 11.43FDD161 pKa = 4.35FFYY164 pKa = 11.19DD165 pKa = 3.01EE166 pKa = 4.34GRR168 pKa = 11.84PYY170 pKa = 9.98TGSYY174 pKa = 5.6WTSSVTRR181 pKa = 11.84IPAEE185 pKa = 3.8NEE187 pKa = 3.72YY188 pKa = 11.34EE189 pKa = 4.16EE190 pKa = 4.35MEE192 pKa = 4.36KK193 pKa = 10.83NYY195 pKa = 10.32GFNWADD201 pKa = 3.35GHH203 pKa = 6.23LKK205 pKa = 10.58SYY207 pKa = 11.16GDD209 pKa = 4.23GYY211 pKa = 11.34SLDD214 pKa = 3.84GSTSGSSIPAGVRR227 pKa = 11.84AVRR230 pKa = 11.84GEE232 pKa = 3.9EE233 pKa = 3.9GVYY236 pKa = 10.46GVNDD240 pKa = 3.48FTDD243 pKa = 3.47NRR245 pKa = 11.84DD246 pKa = 3.29GTITDD251 pKa = 3.46QATGLMWSQKK261 pKa = 10.86DD262 pKa = 3.48SGAVNDD268 pKa = 4.75DD269 pKa = 3.34GTLRR273 pKa = 11.84SEE275 pKa = 4.16DD276 pKa = 4.1DD277 pKa = 3.6EE278 pKa = 4.89NFGLGRR284 pKa = 11.84TWVDD288 pKa = 3.08TLVWVQNMNAAQYY301 pKa = 10.73LGYY304 pKa = 10.16SDD306 pKa = 4.31WRR308 pKa = 11.84LPDD311 pKa = 3.68AKK313 pKa = 9.69EE314 pKa = 3.83LQSIVQYY321 pKa = 10.82GISEE325 pKa = 4.31LPATDD330 pKa = 3.8SDD332 pKa = 4.44YY333 pKa = 11.58FDD335 pKa = 5.05LSRR338 pKa = 11.84TDD340 pKa = 3.08SYY342 pKa = 10.88MWTSTTCGDD351 pKa = 4.09FPDD354 pKa = 3.84TALYY358 pKa = 9.95FAFGKK363 pKa = 10.6AYY365 pKa = 10.12GINLMTGGVPAGQGPEE381 pKa = 4.19GGEE384 pKa = 3.97QMGPPPEE391 pKa = 4.74DD392 pKa = 3.33VNDD395 pKa = 3.7PMEE398 pKa = 4.7NEE400 pKa = 4.07VEE402 pKa = 4.38SFVTVDD408 pKa = 2.97ATVADD413 pKa = 4.82FVDD416 pKa = 3.73THH418 pKa = 6.92GPGAMRR424 pKa = 11.84NDD426 pKa = 3.76YY427 pKa = 11.23KK428 pKa = 11.2DD429 pKa = 3.42VTGTATQAPALSRR442 pKa = 11.84EE443 pKa = 4.39LWSKK447 pKa = 11.12LYY449 pKa = 10.94GEE451 pKa = 5.07EE452 pKa = 4.12YY453 pKa = 9.94PYY455 pKa = 11.44DD456 pKa = 4.34FDD458 pKa = 5.98PSDD461 pKa = 3.51EE462 pKa = 4.25TTLFDD467 pKa = 5.62LSASEE472 pKa = 4.16NAADD476 pKa = 4.82YY477 pKa = 10.82IVLYY481 pKa = 10.7NYY483 pKa = 10.99ALLVRR488 pKa = 11.84DD489 pKa = 3.99ADD491 pKa = 3.7
Molecular weight: 54.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C1QLR6|A0A5C1QLR6_9SPIO PAS domain S-box protein OS=Oceanispirochaeta crateris OX=2518645 GN=EXM22_10740 PE=4 SV=1
MM1 pKa = 7.62AKK3 pKa = 10.23KK4 pKa = 10.87SMIVKK9 pKa = 10.1CNRR12 pKa = 11.84KK13 pKa = 8.78QKK15 pKa = 10.62FSTRR19 pKa = 11.84EE20 pKa = 3.64YY21 pKa = 10.2NRR23 pKa = 11.84CRR25 pKa = 11.84ICGRR29 pKa = 11.84PRR31 pKa = 11.84GYY33 pKa = 8.93MRR35 pKa = 11.84KK36 pKa = 9.48FEE38 pKa = 3.99MCRR41 pKa = 11.84ICFRR45 pKa = 11.84KK46 pKa = 9.58LANEE50 pKa = 3.78GMIPGVTKK58 pKa = 10.78SSWW61 pKa = 2.87
MM1 pKa = 7.62AKK3 pKa = 10.23KK4 pKa = 10.87SMIVKK9 pKa = 10.1CNRR12 pKa = 11.84KK13 pKa = 8.78QKK15 pKa = 10.62FSTRR19 pKa = 11.84EE20 pKa = 3.64YY21 pKa = 10.2NRR23 pKa = 11.84CRR25 pKa = 11.84ICGRR29 pKa = 11.84PRR31 pKa = 11.84GYY33 pKa = 8.93MRR35 pKa = 11.84KK36 pKa = 9.48FEE38 pKa = 3.99MCRR41 pKa = 11.84ICFRR45 pKa = 11.84KK46 pKa = 9.58LANEE50 pKa = 3.78GMIPGVTKK58 pKa = 10.78SSWW61 pKa = 2.87
Molecular weight: 7.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1208583 |
19 |
3847 |
344.9 |
38.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.43 ± 0.042 | 0.979 ± 0.016 |
5.644 ± 0.037 | 7.063 ± 0.046 |
4.677 ± 0.029 | 7.025 ± 0.041 |
1.834 ± 0.02 | 7.692 ± 0.043 |
6.142 ± 0.043 | 10.727 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.828 ± 0.018 | 4.276 ± 0.03 |
3.905 ± 0.024 | 3.336 ± 0.023 |
4.641 ± 0.036 | 7.423 ± 0.035 |
4.925 ± 0.041 | 5.99 ± 0.035 |
1.052 ± 0.013 | 3.411 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
