
Zebra finch circovirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Circovirus
Average proteome isoelectric point is 8.88
Get precalculated fractions of proteins
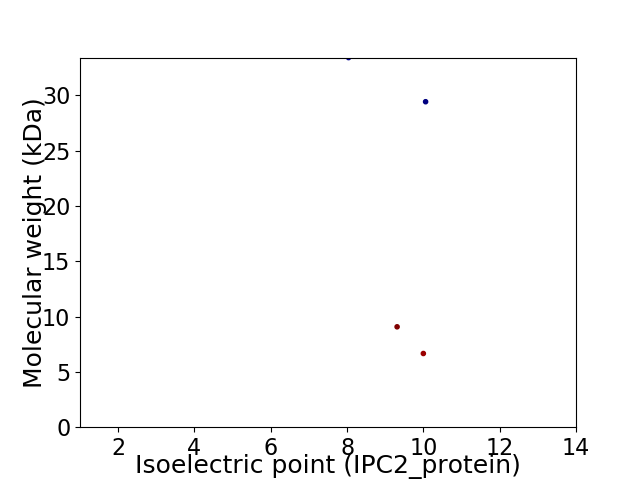
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A142LXX1|A0A142LXX1_9CIRC Capsid protein OS=Zebra finch circovirus OX=1642515 GN=cap PE=3 SV=1
MM1 pKa = 7.84PKK3 pKa = 9.54QARR6 pKa = 11.84EE7 pKa = 4.38SPCKK11 pKa = 10.03RR12 pKa = 11.84WVFTLNNPTEE22 pKa = 4.21EE23 pKa = 3.96EE24 pKa = 4.25VEE26 pKa = 4.26NVKK29 pKa = 10.49KK30 pKa = 10.74LPPSEE35 pKa = 3.77YY36 pKa = 9.92HH37 pKa = 5.22YY38 pKa = 11.01AIVGKK43 pKa = 10.28EE44 pKa = 3.88KK45 pKa = 11.02GEE47 pKa = 3.86QGTPHH52 pKa = 6.35LQGFLHH58 pKa = 6.63LKK60 pKa = 10.07KK61 pKa = 10.27KK62 pKa = 10.34VRR64 pKa = 11.84LNQMKK69 pKa = 10.0QLLPRR74 pKa = 11.84AHH76 pKa = 6.96FEE78 pKa = 3.81RR79 pKa = 11.84ARR81 pKa = 11.84GSDD84 pKa = 3.51EE85 pKa = 6.24DD86 pKa = 3.95NEE88 pKa = 4.6QYY90 pKa = 10.46CSKK93 pKa = 10.32EE94 pKa = 3.49GDD96 pKa = 3.67VILTIGAPAAGNRR109 pKa = 11.84SDD111 pKa = 4.56LAGAVAAVKK120 pKa = 10.28AGRR123 pKa = 11.84AMTEE127 pKa = 3.85VARR130 pKa = 11.84EE131 pKa = 3.7FSEE134 pKa = 6.33AYY136 pKa = 9.91VKK138 pKa = 10.05WGRR141 pKa = 11.84GLKK144 pKa = 10.32DD145 pKa = 3.42LALMIGQKK153 pKa = 10.19PRR155 pKa = 11.84DD156 pKa = 3.87FKK158 pKa = 11.24TEE160 pKa = 4.14VIVLTGPSGVGKK172 pKa = 10.26SRR174 pKa = 11.84WANEE178 pKa = 3.68QEE180 pKa = 4.04GTKK183 pKa = 9.85FYY185 pKa = 11.31KK186 pKa = 10.05MKK188 pKa = 10.64GDD190 pKa = 2.93WWDD193 pKa = 3.88GYY195 pKa = 10.06SNEE198 pKa = 5.05DD199 pKa = 3.24IVVIDD204 pKa = 5.7DD205 pKa = 4.34FYY207 pKa = 11.69GWIPFCEE214 pKa = 4.32LLRR217 pKa = 11.84LTDD220 pKa = 4.49RR221 pKa = 11.84YY222 pKa = 8.43PHH224 pKa = 6.25KK225 pKa = 11.17VPVKK229 pKa = 8.85GSYY232 pKa = 10.43VEE234 pKa = 4.26FTSKK238 pKa = 10.77KK239 pKa = 10.29IIITSNTHH247 pKa = 5.69PDD249 pKa = 2.53HH250 pKa = 6.87WYY252 pKa = 10.6SEE254 pKa = 4.43EE255 pKa = 3.85KK256 pKa = 10.68CYY258 pKa = 10.95VQALFRR264 pKa = 11.84RR265 pKa = 11.84INKK268 pKa = 8.45WLMWDD273 pKa = 3.24VFGFVDD279 pKa = 5.43APDD282 pKa = 3.62AVKK285 pKa = 10.33KK286 pKa = 10.86YY287 pKa = 9.37PINYY291 pKa = 9.36
MM1 pKa = 7.84PKK3 pKa = 9.54QARR6 pKa = 11.84EE7 pKa = 4.38SPCKK11 pKa = 10.03RR12 pKa = 11.84WVFTLNNPTEE22 pKa = 4.21EE23 pKa = 3.96EE24 pKa = 4.25VEE26 pKa = 4.26NVKK29 pKa = 10.49KK30 pKa = 10.74LPPSEE35 pKa = 3.77YY36 pKa = 9.92HH37 pKa = 5.22YY38 pKa = 11.01AIVGKK43 pKa = 10.28EE44 pKa = 3.88KK45 pKa = 11.02GEE47 pKa = 3.86QGTPHH52 pKa = 6.35LQGFLHH58 pKa = 6.63LKK60 pKa = 10.07KK61 pKa = 10.27KK62 pKa = 10.34VRR64 pKa = 11.84LNQMKK69 pKa = 10.0QLLPRR74 pKa = 11.84AHH76 pKa = 6.96FEE78 pKa = 3.81RR79 pKa = 11.84ARR81 pKa = 11.84GSDD84 pKa = 3.51EE85 pKa = 6.24DD86 pKa = 3.95NEE88 pKa = 4.6QYY90 pKa = 10.46CSKK93 pKa = 10.32EE94 pKa = 3.49GDD96 pKa = 3.67VILTIGAPAAGNRR109 pKa = 11.84SDD111 pKa = 4.56LAGAVAAVKK120 pKa = 10.28AGRR123 pKa = 11.84AMTEE127 pKa = 3.85VARR130 pKa = 11.84EE131 pKa = 3.7FSEE134 pKa = 6.33AYY136 pKa = 9.91VKK138 pKa = 10.05WGRR141 pKa = 11.84GLKK144 pKa = 10.32DD145 pKa = 3.42LALMIGQKK153 pKa = 10.19PRR155 pKa = 11.84DD156 pKa = 3.87FKK158 pKa = 11.24TEE160 pKa = 4.14VIVLTGPSGVGKK172 pKa = 10.26SRR174 pKa = 11.84WANEE178 pKa = 3.68QEE180 pKa = 4.04GTKK183 pKa = 9.85FYY185 pKa = 11.31KK186 pKa = 10.05MKK188 pKa = 10.64GDD190 pKa = 2.93WWDD193 pKa = 3.88GYY195 pKa = 10.06SNEE198 pKa = 5.05DD199 pKa = 3.24IVVIDD204 pKa = 5.7DD205 pKa = 4.34FYY207 pKa = 11.69GWIPFCEE214 pKa = 4.32LLRR217 pKa = 11.84LTDD220 pKa = 4.49RR221 pKa = 11.84YY222 pKa = 8.43PHH224 pKa = 6.25KK225 pKa = 11.17VPVKK229 pKa = 8.85GSYY232 pKa = 10.43VEE234 pKa = 4.26FTSKK238 pKa = 10.77KK239 pKa = 10.29IIITSNTHH247 pKa = 5.69PDD249 pKa = 2.53HH250 pKa = 6.87WYY252 pKa = 10.6SEE254 pKa = 4.43EE255 pKa = 3.85KK256 pKa = 10.68CYY258 pKa = 10.95VQALFRR264 pKa = 11.84RR265 pKa = 11.84INKK268 pKa = 8.45WLMWDD273 pKa = 3.24VFGFVDD279 pKa = 5.43APDD282 pKa = 3.62AVKK285 pKa = 10.33KK286 pKa = 10.86YY287 pKa = 9.37PINYY291 pKa = 9.36
Molecular weight: 33.43 kDa
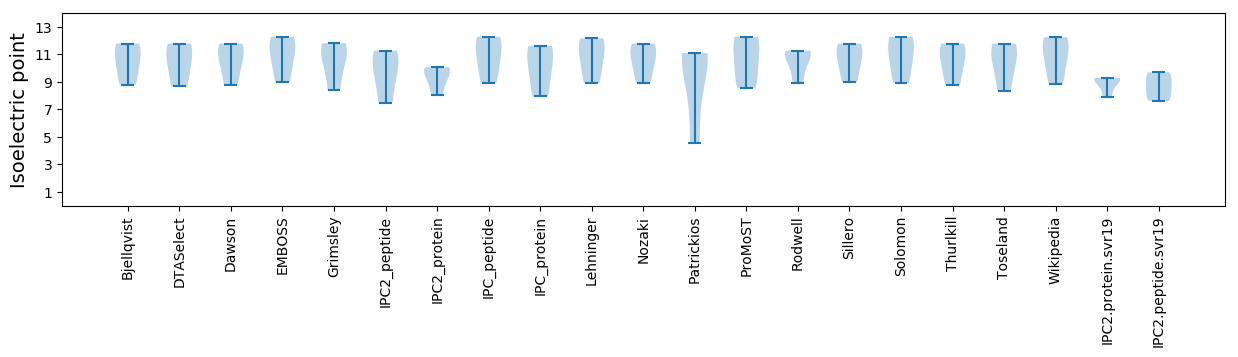
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A142LXX3|A0A142LXX3_9CIRC ORFC4 OS=Zebra finch circovirus OX=1642515 GN=ORFC4 PE=4 SV=1
MM1 pKa = 7.76NEE3 pKa = 3.92YY4 pKa = 10.79CPLRR8 pKa = 11.84TTAFAWPKK16 pKa = 10.18SRR18 pKa = 11.84PSPAMRR24 pKa = 11.84TGRR27 pKa = 11.84TGRR30 pKa = 11.84GRR32 pKa = 11.84ATPSPSKK39 pKa = 8.43THH41 pKa = 4.61TWEE44 pKa = 4.2TSSKK48 pKa = 10.29RR49 pKa = 11.84PSCRR53 pKa = 11.84PTRR56 pKa = 11.84WPIGTEE62 pKa = 4.12PEE64 pKa = 4.13SGTCAKK70 pKa = 10.57ASNAYY75 pKa = 10.03SGPDD79 pKa = 3.26HH80 pKa = 7.0SCPP83 pKa = 3.98
MM1 pKa = 7.76NEE3 pKa = 3.92YY4 pKa = 10.79CPLRR8 pKa = 11.84TTAFAWPKK16 pKa = 10.18SRR18 pKa = 11.84PSPAMRR24 pKa = 11.84TGRR27 pKa = 11.84TGRR30 pKa = 11.84GRR32 pKa = 11.84ATPSPSKK39 pKa = 8.43THH41 pKa = 4.61TWEE44 pKa = 4.2TSSKK48 pKa = 10.29RR49 pKa = 11.84PSCRR53 pKa = 11.84PTRR56 pKa = 11.84WPIGTEE62 pKa = 4.12PEE64 pKa = 4.13SGTCAKK70 pKa = 10.57ASNAYY75 pKa = 10.03SGPDD79 pKa = 3.26HH80 pKa = 7.0SCPP83 pKa = 3.98
Molecular weight: 9.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
680 |
62 |
291 |
170.0 |
19.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.059 ± 0.745 | 1.324 ± 0.811 |
4.265 ± 0.898 | 5.882 ± 1.061 |
3.824 ± 0.727 | 7.5 ± 1.636 |
2.941 ± 0.355 | 3.529 ± 0.676 |
6.324 ± 1.741 | 7.059 ± 1.223 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.206 ± 0.126 | 3.824 ± 0.375 |
7.059 ± 1.564 | 3.235 ± 0.755 |
10.294 ± 2.659 | 5.0 ± 1.745 |
6.912 ± 1.782 | 4.853 ± 1.609 |
3.529 ± 0.381 | 3.382 ± 0.65 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
