
Dorea sp. 5-2
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Dorea; unclassified Dorea
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
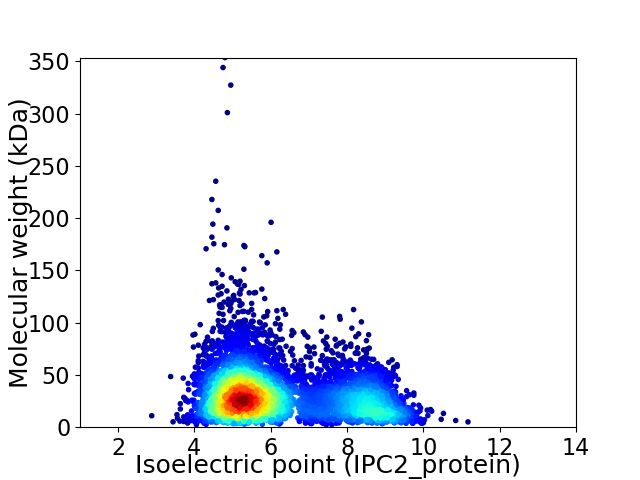
Virtual 2D-PAGE plot for 5824 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R9N8C1|R9N8C1_9FIRM T2SSF domain-containing protein OS=Dorea sp. 5-2 OX=1235798 GN=C817_03003 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 10.08RR3 pKa = 11.84RR4 pKa = 11.84AIAALLTAAMSVSLFTGCHH23 pKa = 6.56DD24 pKa = 4.31SSQEE28 pKa = 3.74TAAVDD33 pKa = 4.46VEE35 pKa = 4.31NTDD38 pKa = 3.46YY39 pKa = 11.47DD40 pKa = 3.66PEE42 pKa = 4.11EE43 pKa = 3.98AVKK46 pKa = 10.74DD47 pKa = 4.12FEE49 pKa = 4.44FGEE52 pKa = 4.19LSEE55 pKa = 4.33EE56 pKa = 4.13EE57 pKa = 3.98KK58 pKa = 11.08NYY60 pKa = 9.03TIEE63 pKa = 4.02MGYY66 pKa = 9.93FNCDD70 pKa = 2.83HH71 pKa = 6.36MVGSIIGDD79 pKa = 3.25KK80 pKa = 10.95AGIYY84 pKa = 7.91EE85 pKa = 4.11ALGLKK90 pKa = 10.52VNVTKK95 pKa = 10.83SSEE98 pKa = 4.16TLKK101 pKa = 11.1ALTSGAMDD109 pKa = 3.35VGYY112 pKa = 8.94TGVTGAVRR120 pKa = 11.84AVNQGAPFFIAAANHH135 pKa = 6.1MGGSMYY141 pKa = 10.75LVASNEE147 pKa = 3.89IKK149 pKa = 9.72TAEE152 pKa = 4.07DD153 pKa = 3.51LKK155 pKa = 11.37GKK157 pKa = 7.27TLSITATPDD166 pKa = 2.92IDD168 pKa = 3.78PTILTWEE175 pKa = 4.16QEE177 pKa = 3.99IGMSSNPEE185 pKa = 3.83DD186 pKa = 4.07YY187 pKa = 10.74EE188 pKa = 4.17IVDD191 pKa = 4.01MGQQDD196 pKa = 3.71AMFALKK202 pKa = 10.57AEE204 pKa = 4.34QIDD207 pKa = 4.16AFSCCDD213 pKa = 3.64PYY215 pKa = 11.86ASIAEE220 pKa = 4.13FEE222 pKa = 4.56GFGHH226 pKa = 6.96IMAIDD231 pKa = 3.85WAAPDD236 pKa = 3.5ISSDD240 pKa = 4.42SEE242 pKa = 4.02YY243 pKa = 11.2DD244 pKa = 3.81DD245 pKa = 4.53WGLCCVYY252 pKa = 11.27AMNTDD257 pKa = 4.48FSKK260 pKa = 11.01NCPEE264 pKa = 3.98LARR267 pKa = 11.84RR268 pKa = 11.84LIFAHH273 pKa = 6.28SLAIKK278 pKa = 10.26YY279 pKa = 8.91MYY281 pKa = 8.51EE282 pKa = 3.98HH283 pKa = 7.8PYY285 pKa = 8.78NAAMMFADD293 pKa = 5.07GFDD296 pKa = 3.48VDD298 pKa = 4.54PYY300 pKa = 10.18VALRR304 pKa = 11.84TIYY307 pKa = 9.87MKK309 pKa = 10.15TVSEE313 pKa = 4.19GRR315 pKa = 11.84TLTWKK320 pKa = 10.75FSDD323 pKa = 3.86PNVDD327 pKa = 4.16SFLEE331 pKa = 4.55YY332 pKa = 9.14YY333 pKa = 8.36TQYY336 pKa = 11.01SQIPEE341 pKa = 4.11EE342 pKa = 4.9EE343 pKa = 3.89IPLINDD349 pKa = 3.32RR350 pKa = 11.84NTLISSEE357 pKa = 4.1VLEE360 pKa = 4.29AAEE363 pKa = 4.48IEE365 pKa = 4.64DD366 pKa = 3.74FQTYY370 pKa = 9.99IKK372 pKa = 10.86DD373 pKa = 3.67VVDD376 pKa = 4.89PIMPLGTTFEE386 pKa = 4.15DD387 pKa = 3.62WYY389 pKa = 10.02EE390 pKa = 3.73ISKK393 pKa = 10.69EE394 pKa = 3.47IDD396 pKa = 4.02GISDD400 pKa = 3.31EE401 pKa = 4.51DD402 pKa = 3.86AVDD405 pKa = 3.59ISDD408 pKa = 3.62TATPYY413 pKa = 11.09LNEE416 pKa = 4.23NLDD419 pKa = 3.71EE420 pKa = 4.35RR421 pKa = 11.84TSNN424 pKa = 3.44
MM1 pKa = 7.44KK2 pKa = 10.08RR3 pKa = 11.84RR4 pKa = 11.84AIAALLTAAMSVSLFTGCHH23 pKa = 6.56DD24 pKa = 4.31SSQEE28 pKa = 3.74TAAVDD33 pKa = 4.46VEE35 pKa = 4.31NTDD38 pKa = 3.46YY39 pKa = 11.47DD40 pKa = 3.66PEE42 pKa = 4.11EE43 pKa = 3.98AVKK46 pKa = 10.74DD47 pKa = 4.12FEE49 pKa = 4.44FGEE52 pKa = 4.19LSEE55 pKa = 4.33EE56 pKa = 4.13EE57 pKa = 3.98KK58 pKa = 11.08NYY60 pKa = 9.03TIEE63 pKa = 4.02MGYY66 pKa = 9.93FNCDD70 pKa = 2.83HH71 pKa = 6.36MVGSIIGDD79 pKa = 3.25KK80 pKa = 10.95AGIYY84 pKa = 7.91EE85 pKa = 4.11ALGLKK90 pKa = 10.52VNVTKK95 pKa = 10.83SSEE98 pKa = 4.16TLKK101 pKa = 11.1ALTSGAMDD109 pKa = 3.35VGYY112 pKa = 8.94TGVTGAVRR120 pKa = 11.84AVNQGAPFFIAAANHH135 pKa = 6.1MGGSMYY141 pKa = 10.75LVASNEE147 pKa = 3.89IKK149 pKa = 9.72TAEE152 pKa = 4.07DD153 pKa = 3.51LKK155 pKa = 11.37GKK157 pKa = 7.27TLSITATPDD166 pKa = 2.92IDD168 pKa = 3.78PTILTWEE175 pKa = 4.16QEE177 pKa = 3.99IGMSSNPEE185 pKa = 3.83DD186 pKa = 4.07YY187 pKa = 10.74EE188 pKa = 4.17IVDD191 pKa = 4.01MGQQDD196 pKa = 3.71AMFALKK202 pKa = 10.57AEE204 pKa = 4.34QIDD207 pKa = 4.16AFSCCDD213 pKa = 3.64PYY215 pKa = 11.86ASIAEE220 pKa = 4.13FEE222 pKa = 4.56GFGHH226 pKa = 6.96IMAIDD231 pKa = 3.85WAAPDD236 pKa = 3.5ISSDD240 pKa = 4.42SEE242 pKa = 4.02YY243 pKa = 11.2DD244 pKa = 3.81DD245 pKa = 4.53WGLCCVYY252 pKa = 11.27AMNTDD257 pKa = 4.48FSKK260 pKa = 11.01NCPEE264 pKa = 3.98LARR267 pKa = 11.84RR268 pKa = 11.84LIFAHH273 pKa = 6.28SLAIKK278 pKa = 10.26YY279 pKa = 8.91MYY281 pKa = 8.51EE282 pKa = 3.98HH283 pKa = 7.8PYY285 pKa = 8.78NAAMMFADD293 pKa = 5.07GFDD296 pKa = 3.48VDD298 pKa = 4.54PYY300 pKa = 10.18VALRR304 pKa = 11.84TIYY307 pKa = 9.87MKK309 pKa = 10.15TVSEE313 pKa = 4.19GRR315 pKa = 11.84TLTWKK320 pKa = 10.75FSDD323 pKa = 3.86PNVDD327 pKa = 4.16SFLEE331 pKa = 4.55YY332 pKa = 9.14YY333 pKa = 8.36TQYY336 pKa = 11.01SQIPEE341 pKa = 4.11EE342 pKa = 4.9EE343 pKa = 3.89IPLINDD349 pKa = 3.32RR350 pKa = 11.84NTLISSEE357 pKa = 4.1VLEE360 pKa = 4.29AAEE363 pKa = 4.48IEE365 pKa = 4.64DD366 pKa = 3.74FQTYY370 pKa = 9.99IKK372 pKa = 10.86DD373 pKa = 3.67VVDD376 pKa = 4.89PIMPLGTTFEE386 pKa = 4.15DD387 pKa = 3.62WYY389 pKa = 10.02EE390 pKa = 3.73ISKK393 pKa = 10.69EE394 pKa = 3.47IDD396 pKa = 4.02GISDD400 pKa = 3.31EE401 pKa = 4.51DD402 pKa = 3.86AVDD405 pKa = 3.59ISDD408 pKa = 3.62TATPYY413 pKa = 11.09LNEE416 pKa = 4.23NLDD419 pKa = 3.71EE420 pKa = 4.35RR421 pKa = 11.84TSNN424 pKa = 3.44
Molecular weight: 47.06 kDa
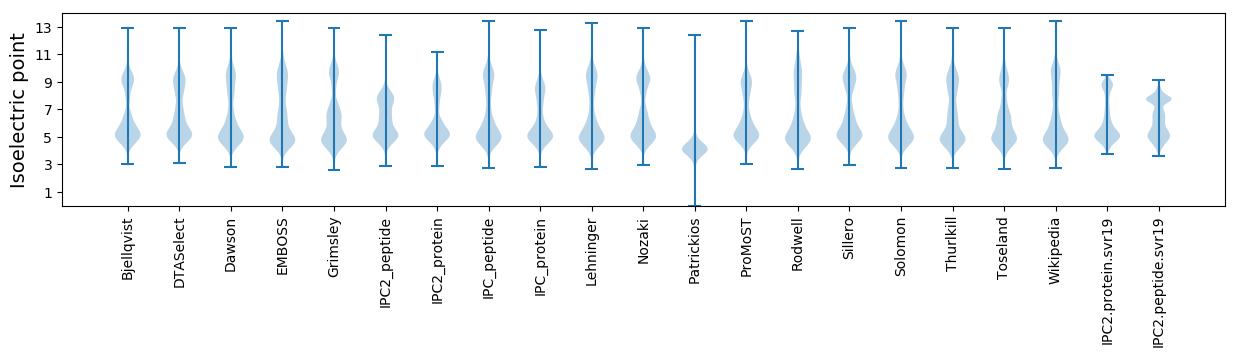
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R9MKG5|R9MKG5_9FIRM Uncharacterized protein OS=Dorea sp. 5-2 OX=1235798 GN=C817_05762 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MSSAGGRR28 pKa = 11.84KK29 pKa = 8.88VLANRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.85GRR39 pKa = 11.84KK40 pKa = 8.87RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MSSAGGRR28 pKa = 11.84KK29 pKa = 8.88VLANRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.85GRR39 pKa = 11.84KK40 pKa = 8.87RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1704686 |
21 |
3121 |
292.7 |
33.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.372 ± 0.033 | 1.569 ± 0.013 |
5.625 ± 0.027 | 8.033 ± 0.042 |
4.035 ± 0.021 | 7.072 ± 0.032 |
1.804 ± 0.014 | 7.199 ± 0.03 |
6.729 ± 0.028 | 8.958 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.164 ± 0.017 | 4.09 ± 0.021 |
3.25 ± 0.019 | 3.349 ± 0.018 |
5.105 ± 0.03 | 5.711 ± 0.026 |
5.051 ± 0.023 | 6.604 ± 0.023 |
0.977 ± 0.011 | 4.306 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
