
Vogesella sp. LIG4
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Neisseriales; Chromobacteriaceae; Vogesella; unclassified Vogesella
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
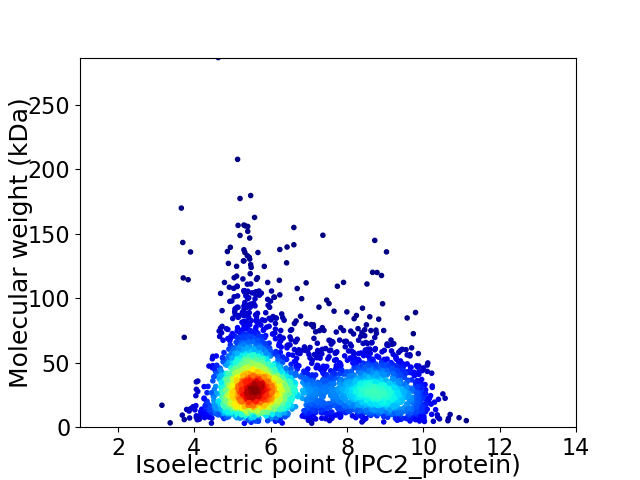
Virtual 2D-PAGE plot for 3722 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
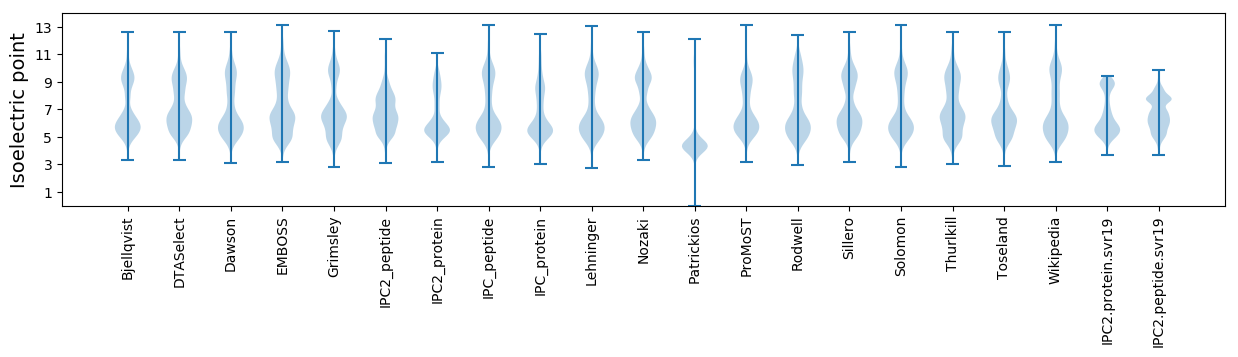
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1C6MT89|A0A1C6MT89_9NEIS Acyl-CoA reductase OS=Vogesella sp. LIG4 OX=1192162 GN=PSELUDRAFT_2512 PE=3 SV=1
SS1 pKa = 7.15ASVDD5 pKa = 3.56ADD7 pKa = 4.01GTWHH11 pKa = 5.47YY12 pKa = 9.31TVSDD16 pKa = 3.86SGTVDD21 pKa = 3.17ALAAGEE27 pKa = 4.13TLGDD31 pKa = 3.79SFTVRR36 pKa = 11.84VSDD39 pKa = 3.43NHH41 pKa = 6.89GGFVDD46 pKa = 3.6QLVSVTITGTNDD58 pKa = 2.83APLIDD63 pKa = 4.54AANTTASGGVSEE75 pKa = 5.68GDD77 pKa = 3.8DD78 pKa = 3.28NSSRR82 pKa = 11.84STSGVIAYY90 pKa = 10.36SDD92 pKa = 3.39VDD94 pKa = 3.5ASDD97 pKa = 3.35SHH99 pKa = 8.18SFALQGTAAAYY110 pKa = 7.44GTASVDD116 pKa = 4.64ADD118 pKa = 4.13GTWHH122 pKa = 5.39YY123 pKa = 9.39TVHH126 pKa = 7.81DD127 pKa = 4.25SGAVDD132 pKa = 3.38ALGAGEE138 pKa = 4.16TLGDD142 pKa = 3.86SFTVRR147 pKa = 11.84VSDD150 pKa = 3.44NHH152 pKa = 6.15GGYY155 pKa = 10.01VDD157 pKa = 3.66QLVSVTITGTNDD169 pKa = 2.88AAVITGTSSASLTEE183 pKa = 4.24SNAAQSTSGTLSISDD198 pKa = 3.55VDD200 pKa = 4.19SPASFVVQNNAAGSNGYY217 pKa = 10.47GKK219 pKa = 10.55FSIDD223 pKa = 3.99AAGHH227 pKa = 5.67WSYY230 pKa = 11.09TMNSAHH236 pKa = 7.23DD237 pKa = 3.95EE238 pKa = 4.41FVGGTTYY245 pKa = 10.77TDD247 pKa = 3.89SITVSSADD255 pKa = 3.19GTTQLLTVSILGTNDD270 pKa = 2.97AAVITGTSSASLTEE284 pKa = 4.35SNVAQSTSGTLSISDD299 pKa = 3.55VDD301 pKa = 4.22SPASFVAQNNVAGSNGYY318 pKa = 10.36GKK320 pKa = 10.55FSIDD324 pKa = 4.03SAGHH328 pKa = 5.54WSYY331 pKa = 11.15TMNSAHH337 pKa = 7.23DD338 pKa = 3.95EE339 pKa = 4.41FVGGTTYY346 pKa = 11.19TDD348 pKa = 3.28TLTVSSADD356 pKa = 3.16GTTQLLTVNILGTNDD371 pKa = 3.18AAVITGTSSTSLTEE385 pKa = 4.2SNVAQSTSGTLSISDD400 pKa = 3.55VDD402 pKa = 4.19SPASFVVQTNVDD414 pKa = 3.67GNHH417 pKa = 6.92GYY419 pKa = 11.38GKK421 pKa = 10.44FSIDD425 pKa = 3.99AAGHH429 pKa = 5.67WSYY432 pKa = 11.09TMNSAHH438 pKa = 7.23DD439 pKa = 3.95EE440 pKa = 4.41FVGGTTYY447 pKa = 11.19TDD449 pKa = 3.28TLTVSSADD457 pKa = 3.16GTTQLLTVSILGTNDD472 pKa = 2.97AAVITGTSSASLTEE486 pKa = 4.35SNVAQSTSGTLSISDD501 pKa = 3.55VDD503 pKa = 4.22SPASFVAQSNAAGSNGYY520 pKa = 10.47GKK522 pKa = 10.55FSIDD526 pKa = 3.99AAGHH530 pKa = 5.67WSYY533 pKa = 11.09TMNSAHH539 pKa = 7.23DD540 pKa = 3.95EE541 pKa = 4.41FVGGTTYY548 pKa = 10.77TDD550 pKa = 3.89SITVSSADD558 pKa = 3.19GTTQLLTVSILGTNDD573 pKa = 2.97AAVITGTSTAALTEE587 pKa = 4.61SNVAQSTSGTLSISDD602 pKa = 3.55VDD604 pKa = 4.19SPASFVVQSNVAGSNGYY621 pKa = 10.51GKK623 pKa = 10.55FSIDD627 pKa = 3.99AAGHH631 pKa = 5.67WSYY634 pKa = 11.09TMNSAHH640 pKa = 7.23DD641 pKa = 3.95EE642 pKa = 4.41FVGGTTYY649 pKa = 10.77TDD651 pKa = 3.89SITVSSADD659 pKa = 3.19GTTQLLTVSILGTNDD674 pKa = 3.76APVLDD679 pKa = 5.0LNGASAGSDD688 pKa = 3.09TTASYY693 pKa = 7.67PTGVSQILIAPAATVTDD710 pKa = 3.48IDD712 pKa = 4.13SANLQSMTVTLTNSPDD728 pKa = 3.59SSSGAARR735 pKa = 11.84EE736 pKa = 3.97ILSLNATATAAAAGLTVSGVSSSNNNVYY764 pKa = 8.86TLTISGAASVATYY777 pKa = 10.29QSILDD782 pKa = 3.76GVQYY786 pKa = 11.17TDD788 pKa = 3.56IKK790 pKa = 9.73TGNHH794 pKa = 3.92NTAARR799 pKa = 11.84TVNVVVSDD807 pKa = 4.34GSASSVLHH815 pKa = 5.66TVTISVAAPAGVAGTPISLAIADD838 pKa = 4.52PSLGTHH844 pKa = 6.87DD845 pKa = 4.51GVSTTITGVPSDD857 pKa = 3.71WSIDD861 pKa = 3.62GATKK865 pKa = 10.27NANGSWTAYY874 pKa = 9.71ISDD877 pKa = 3.92PGMLTVTTPSGFAGAAVLTVTQTWVNADD905 pKa = 3.22GSEE908 pKa = 3.81GRR910 pKa = 11.84AIILDD915 pKa = 3.68NVEE918 pKa = 4.59AYY920 pKa = 10.49APGSPIFALSSDD932 pKa = 4.12DD933 pKa = 3.95NLTGSSAADD942 pKa = 3.14TFVFAQPMGNNMIRR956 pKa = 11.84NFNPANDD963 pKa = 4.54KK964 pKa = 10.63IDD966 pKa = 4.81LIGFAGVGSIANLQLANDD984 pKa = 4.0ANGNAVISIGNGQSITLLGVDD1005 pKa = 4.19VSLLLAANFLFNQVPVLHH1023 pKa = 7.09NDD1025 pKa = 3.14GVMTIADD1032 pKa = 4.12GAILPLGGEE1041 pKa = 4.28VDD1043 pKa = 3.54NSGSIVLASSGGATSLEE1060 pKa = 4.1VLVDD1064 pKa = 3.8SVTLQGGGKK1073 pKa = 9.93LLLSDD1078 pKa = 5.18DD1079 pKa = 3.76SHH1081 pKa = 8.14NSVFGGAGSATLVNVDD1097 pKa = 3.7NTISGAGQLGGGQMTLLNHH1116 pKa = 6.42GSIVADD1122 pKa = 4.28GSHH1125 pKa = 7.09ALLLDD1130 pKa = 3.37TGGNSIVNSGQLHH1143 pKa = 6.2ASGAGGMVVASSLEE1157 pKa = 3.85NSGNLWADD1165 pKa = 3.24NSSLQLQQDD1174 pKa = 3.55VSGQGSALISGNGSIAFMAAADD1196 pKa = 3.99TDD1198 pKa = 3.82VSFANGSHH1206 pKa = 6.83GDD1208 pKa = 3.82LLLDD1212 pKa = 4.08HH1213 pKa = 7.48ADD1215 pKa = 3.47AFSGRR1220 pKa = 11.84IIGMASGDD1228 pKa = 3.88HH1229 pKa = 6.87LDD1231 pKa = 4.75LGGLAYY1237 pKa = 10.74NDD1239 pKa = 4.24GLRR1242 pKa = 11.84LDD1244 pKa = 4.57YY1245 pKa = 11.01IGDD1248 pKa = 3.62SSGIGGKK1255 pKa = 9.84LLIINGSDD1263 pKa = 3.3TTSLGLIGHH1272 pKa = 5.86YY1273 pKa = 9.27QASDD1277 pKa = 3.57FQLAAAADD1285 pKa = 3.78GHH1287 pKa = 5.97SVVNTSQTDD1296 pKa = 3.15SSTVIGTAGDD1306 pKa = 3.73DD1307 pKa = 3.82TLQGSSGNDD1316 pKa = 2.83ILLAGAGHH1324 pKa = 6.0NTLTGGGGSDD1334 pKa = 3.32TYY1336 pKa = 11.81VLLRR1340 pKa = 11.84SDD1342 pKa = 3.92GNAVDD1347 pKa = 4.87TITDD1351 pKa = 4.19FKK1353 pKa = 11.1PGDD1356 pKa = 4.16GGDD1359 pKa = 3.51KK1360 pKa = 10.83LQIGDD1365 pKa = 4.95LLTGYY1370 pKa = 10.31QPDD1373 pKa = 3.67AANQFLSLRR1382 pKa = 11.84EE1383 pKa = 4.09EE1384 pKa = 4.3NGNTVISVDD1393 pKa = 3.41RR1394 pKa = 11.84DD1395 pKa = 3.58GAGGKK1400 pKa = 9.09YY1401 pKa = 9.12QMQDD1405 pKa = 2.89MVVLQGVTGLDD1416 pKa = 3.22LTTLLKK1422 pKa = 10.77HH1423 pKa = 6.39VDD1425 pKa = 3.2TSQII1429 pKa = 3.31
SS1 pKa = 7.15ASVDD5 pKa = 3.56ADD7 pKa = 4.01GTWHH11 pKa = 5.47YY12 pKa = 9.31TVSDD16 pKa = 3.86SGTVDD21 pKa = 3.17ALAAGEE27 pKa = 4.13TLGDD31 pKa = 3.79SFTVRR36 pKa = 11.84VSDD39 pKa = 3.43NHH41 pKa = 6.89GGFVDD46 pKa = 3.6QLVSVTITGTNDD58 pKa = 2.83APLIDD63 pKa = 4.54AANTTASGGVSEE75 pKa = 5.68GDD77 pKa = 3.8DD78 pKa = 3.28NSSRR82 pKa = 11.84STSGVIAYY90 pKa = 10.36SDD92 pKa = 3.39VDD94 pKa = 3.5ASDD97 pKa = 3.35SHH99 pKa = 8.18SFALQGTAAAYY110 pKa = 7.44GTASVDD116 pKa = 4.64ADD118 pKa = 4.13GTWHH122 pKa = 5.39YY123 pKa = 9.39TVHH126 pKa = 7.81DD127 pKa = 4.25SGAVDD132 pKa = 3.38ALGAGEE138 pKa = 4.16TLGDD142 pKa = 3.86SFTVRR147 pKa = 11.84VSDD150 pKa = 3.44NHH152 pKa = 6.15GGYY155 pKa = 10.01VDD157 pKa = 3.66QLVSVTITGTNDD169 pKa = 2.88AAVITGTSSASLTEE183 pKa = 4.24SNAAQSTSGTLSISDD198 pKa = 3.55VDD200 pKa = 4.19SPASFVVQNNAAGSNGYY217 pKa = 10.47GKK219 pKa = 10.55FSIDD223 pKa = 3.99AAGHH227 pKa = 5.67WSYY230 pKa = 11.09TMNSAHH236 pKa = 7.23DD237 pKa = 3.95EE238 pKa = 4.41FVGGTTYY245 pKa = 10.77TDD247 pKa = 3.89SITVSSADD255 pKa = 3.19GTTQLLTVSILGTNDD270 pKa = 2.97AAVITGTSSASLTEE284 pKa = 4.35SNVAQSTSGTLSISDD299 pKa = 3.55VDD301 pKa = 4.22SPASFVAQNNVAGSNGYY318 pKa = 10.36GKK320 pKa = 10.55FSIDD324 pKa = 4.03SAGHH328 pKa = 5.54WSYY331 pKa = 11.15TMNSAHH337 pKa = 7.23DD338 pKa = 3.95EE339 pKa = 4.41FVGGTTYY346 pKa = 11.19TDD348 pKa = 3.28TLTVSSADD356 pKa = 3.16GTTQLLTVNILGTNDD371 pKa = 3.18AAVITGTSSTSLTEE385 pKa = 4.2SNVAQSTSGTLSISDD400 pKa = 3.55VDD402 pKa = 4.19SPASFVVQTNVDD414 pKa = 3.67GNHH417 pKa = 6.92GYY419 pKa = 11.38GKK421 pKa = 10.44FSIDD425 pKa = 3.99AAGHH429 pKa = 5.67WSYY432 pKa = 11.09TMNSAHH438 pKa = 7.23DD439 pKa = 3.95EE440 pKa = 4.41FVGGTTYY447 pKa = 11.19TDD449 pKa = 3.28TLTVSSADD457 pKa = 3.16GTTQLLTVSILGTNDD472 pKa = 2.97AAVITGTSSASLTEE486 pKa = 4.35SNVAQSTSGTLSISDD501 pKa = 3.55VDD503 pKa = 4.22SPASFVAQSNAAGSNGYY520 pKa = 10.47GKK522 pKa = 10.55FSIDD526 pKa = 3.99AAGHH530 pKa = 5.67WSYY533 pKa = 11.09TMNSAHH539 pKa = 7.23DD540 pKa = 3.95EE541 pKa = 4.41FVGGTTYY548 pKa = 10.77TDD550 pKa = 3.89SITVSSADD558 pKa = 3.19GTTQLLTVSILGTNDD573 pKa = 2.97AAVITGTSTAALTEE587 pKa = 4.61SNVAQSTSGTLSISDD602 pKa = 3.55VDD604 pKa = 4.19SPASFVVQSNVAGSNGYY621 pKa = 10.51GKK623 pKa = 10.55FSIDD627 pKa = 3.99AAGHH631 pKa = 5.67WSYY634 pKa = 11.09TMNSAHH640 pKa = 7.23DD641 pKa = 3.95EE642 pKa = 4.41FVGGTTYY649 pKa = 10.77TDD651 pKa = 3.89SITVSSADD659 pKa = 3.19GTTQLLTVSILGTNDD674 pKa = 3.76APVLDD679 pKa = 5.0LNGASAGSDD688 pKa = 3.09TTASYY693 pKa = 7.67PTGVSQILIAPAATVTDD710 pKa = 3.48IDD712 pKa = 4.13SANLQSMTVTLTNSPDD728 pKa = 3.59SSSGAARR735 pKa = 11.84EE736 pKa = 3.97ILSLNATATAAAAGLTVSGVSSSNNNVYY764 pKa = 8.86TLTISGAASVATYY777 pKa = 10.29QSILDD782 pKa = 3.76GVQYY786 pKa = 11.17TDD788 pKa = 3.56IKK790 pKa = 9.73TGNHH794 pKa = 3.92NTAARR799 pKa = 11.84TVNVVVSDD807 pKa = 4.34GSASSVLHH815 pKa = 5.66TVTISVAAPAGVAGTPISLAIADD838 pKa = 4.52PSLGTHH844 pKa = 6.87DD845 pKa = 4.51GVSTTITGVPSDD857 pKa = 3.71WSIDD861 pKa = 3.62GATKK865 pKa = 10.27NANGSWTAYY874 pKa = 9.71ISDD877 pKa = 3.92PGMLTVTTPSGFAGAAVLTVTQTWVNADD905 pKa = 3.22GSEE908 pKa = 3.81GRR910 pKa = 11.84AIILDD915 pKa = 3.68NVEE918 pKa = 4.59AYY920 pKa = 10.49APGSPIFALSSDD932 pKa = 4.12DD933 pKa = 3.95NLTGSSAADD942 pKa = 3.14TFVFAQPMGNNMIRR956 pKa = 11.84NFNPANDD963 pKa = 4.54KK964 pKa = 10.63IDD966 pKa = 4.81LIGFAGVGSIANLQLANDD984 pKa = 4.0ANGNAVISIGNGQSITLLGVDD1005 pKa = 4.19VSLLLAANFLFNQVPVLHH1023 pKa = 7.09NDD1025 pKa = 3.14GVMTIADD1032 pKa = 4.12GAILPLGGEE1041 pKa = 4.28VDD1043 pKa = 3.54NSGSIVLASSGGATSLEE1060 pKa = 4.1VLVDD1064 pKa = 3.8SVTLQGGGKK1073 pKa = 9.93LLLSDD1078 pKa = 5.18DD1079 pKa = 3.76SHH1081 pKa = 8.14NSVFGGAGSATLVNVDD1097 pKa = 3.7NTISGAGQLGGGQMTLLNHH1116 pKa = 6.42GSIVADD1122 pKa = 4.28GSHH1125 pKa = 7.09ALLLDD1130 pKa = 3.37TGGNSIVNSGQLHH1143 pKa = 6.2ASGAGGMVVASSLEE1157 pKa = 3.85NSGNLWADD1165 pKa = 3.24NSSLQLQQDD1174 pKa = 3.55VSGQGSALISGNGSIAFMAAADD1196 pKa = 3.99TDD1198 pKa = 3.82VSFANGSHH1206 pKa = 6.83GDD1208 pKa = 3.82LLLDD1212 pKa = 4.08HH1213 pKa = 7.48ADD1215 pKa = 3.47AFSGRR1220 pKa = 11.84IIGMASGDD1228 pKa = 3.88HH1229 pKa = 6.87LDD1231 pKa = 4.75LGGLAYY1237 pKa = 10.74NDD1239 pKa = 4.24GLRR1242 pKa = 11.84LDD1244 pKa = 4.57YY1245 pKa = 11.01IGDD1248 pKa = 3.62SSGIGGKK1255 pKa = 9.84LLIINGSDD1263 pKa = 3.3TTSLGLIGHH1272 pKa = 5.86YY1273 pKa = 9.27QASDD1277 pKa = 3.57FQLAAAADD1285 pKa = 3.78GHH1287 pKa = 5.97SVVNTSQTDD1296 pKa = 3.15SSTVIGTAGDD1306 pKa = 3.73DD1307 pKa = 3.82TLQGSSGNDD1316 pKa = 2.83ILLAGAGHH1324 pKa = 6.0NTLTGGGGSDD1334 pKa = 3.32TYY1336 pKa = 11.81VLLRR1340 pKa = 11.84SDD1342 pKa = 3.92GNAVDD1347 pKa = 4.87TITDD1351 pKa = 4.19FKK1353 pKa = 11.1PGDD1356 pKa = 4.16GGDD1359 pKa = 3.51KK1360 pKa = 10.83LQIGDD1365 pKa = 4.95LLTGYY1370 pKa = 10.31QPDD1373 pKa = 3.67AANQFLSLRR1382 pKa = 11.84EE1383 pKa = 4.09EE1384 pKa = 4.3NGNTVISVDD1393 pKa = 3.41RR1394 pKa = 11.84DD1395 pKa = 3.58GAGGKK1400 pKa = 9.09YY1401 pKa = 9.12QMQDD1405 pKa = 2.89MVVLQGVTGLDD1416 pKa = 3.22LTTLLKK1422 pKa = 10.77HH1423 pKa = 6.39VDD1425 pKa = 3.2TSQII1429 pKa = 3.31
Molecular weight: 143.11 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1C6M0F0|A0A1C6M0F0_9NEIS Uncharacterized protein OS=Vogesella sp. LIG4 OX=1192162 GN=PSELUDRAFT_1090 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.29QPSNTRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.05RR14 pKa = 11.84THH16 pKa = 5.88GFLVRR21 pKa = 11.84SKK23 pKa = 9.38TRR25 pKa = 11.84GGRR28 pKa = 11.84AVLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.87GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.29QPSNTRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.05RR14 pKa = 11.84THH16 pKa = 5.88GFLVRR21 pKa = 11.84SKK23 pKa = 9.38TRR25 pKa = 11.84GGRR28 pKa = 11.84AVLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.87GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1207113 |
26 |
2768 |
324.3 |
35.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.146 ± 0.059 | 1.01 ± 0.015 |
5.274 ± 0.028 | 5.245 ± 0.038 |
3.5 ± 0.021 | 7.984 ± 0.039 |
2.404 ± 0.021 | 4.618 ± 0.034 |
3.338 ± 0.04 | 11.79 ± 0.064 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.468 ± 0.019 | 2.892 ± 0.027 |
4.878 ± 0.034 | 4.799 ± 0.039 |
6.358 ± 0.039 | 5.626 ± 0.037 |
4.627 ± 0.033 | 7.122 ± 0.038 |
1.402 ± 0.016 | 2.519 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
