
Sphingomonas parva
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingomonas
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
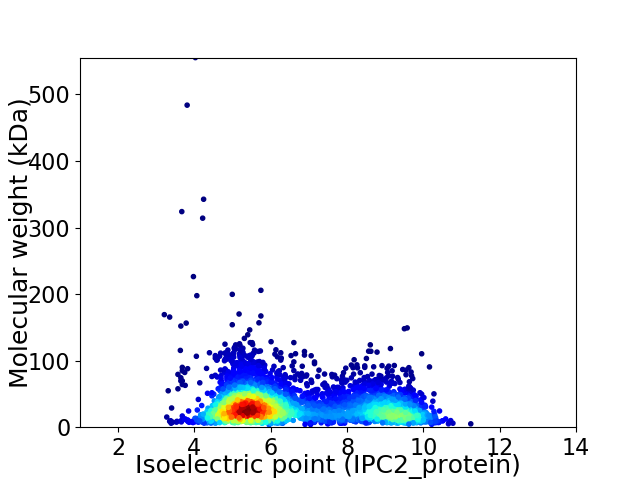
Virtual 2D-PAGE plot for 4021 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y8ZKF2|A0A4Y8ZKF2_9SPHN NADPH-dependent 7-cyano-7-deazaguanine reductase QueF (Fragment) OS=Sphingomonas parva OX=2555898 GN=E2493_20045 PE=4 SV=1
MM1 pKa = 7.38GGSRR5 pKa = 11.84MSTEE9 pKa = 4.38LGFDD13 pKa = 3.34APASNLVPLEE23 pKa = 4.67EE24 pKa = 4.28IAHH27 pKa = 4.77TTFVEE32 pKa = 4.87GCGCAACQGFQDD44 pKa = 4.08AGTDD48 pKa = 3.94GVISKK53 pKa = 10.96GGFNEE58 pKa = 4.2ANPEE62 pKa = 4.04SGGIGVLLPLVTNPDD77 pKa = 3.23GSRR80 pKa = 11.84SITGEE85 pKa = 3.97RR86 pKa = 11.84NVDD89 pKa = 3.97AILIGAKK96 pKa = 9.12WGATNLTFSFPKK108 pKa = 10.42SGSNYY113 pKa = 9.86NGTGFDD119 pKa = 3.86SNGVSIYY126 pKa = 10.77HH127 pKa = 6.11MEE129 pKa = 5.19LGAQQQAAARR139 pKa = 11.84AAFAQISAATGLTFTEE155 pKa = 4.14ITEE158 pKa = 4.3TDD160 pKa = 3.41TVHH163 pKa = 6.72ANIRR167 pKa = 11.84ISQSGDD173 pKa = 2.96NDD175 pKa = 3.36VTSAYY180 pKa = 10.54GGFPSDD186 pKa = 3.34TRR188 pKa = 11.84GVAGDD193 pKa = 2.84IWFGRR198 pKa = 11.84TSQPYY203 pKa = 9.8YY204 pKa = 10.86DD205 pKa = 4.37LAYY208 pKa = 10.03KK209 pKa = 7.78GTWGYY214 pKa = 8.84ATMMHH219 pKa = 7.03EE220 pKa = 4.66IGHH223 pKa = 5.19TMGLKK228 pKa = 10.05HH229 pKa = 6.31GHH231 pKa = 5.58QDD233 pKa = 3.44YY234 pKa = 9.86TNSDD238 pKa = 3.12LSFYY242 pKa = 10.68FGTSPRR248 pKa = 11.84RR249 pKa = 11.84GTQSLTPDD257 pKa = 3.43NDD259 pKa = 3.47GQAWSLMTYY268 pKa = 9.37TPAPFTNSNFAGEE281 pKa = 4.87KK282 pKa = 9.42VNQPQTYY289 pKa = 7.5MQYY292 pKa = 11.57DD293 pKa = 4.48LAALQYY299 pKa = 10.17MYY301 pKa = 10.94GANYY305 pKa = 7.39NTNAGDD311 pKa = 3.89TVYY314 pKa = 9.39TWSEE318 pKa = 3.99TTGEE322 pKa = 4.12MFVNGVGQGAPSGNTILLTIWDD344 pKa = 4.15GGGVDD349 pKa = 4.76TLDD352 pKa = 3.56LSNYY356 pKa = 9.71ADD358 pKa = 3.77GVTVDD363 pKa = 4.27LRR365 pKa = 11.84PGQFSTFDD373 pKa = 3.24QDD375 pKa = 3.52QLANHH380 pKa = 7.39LAYY383 pKa = 10.65QNLTALAPGNVAMSLLYY400 pKa = 11.17NNDD403 pKa = 2.66GRR405 pKa = 11.84SLIEE409 pKa = 3.83NAKK412 pKa = 10.49GGIGNDD418 pKa = 3.17VFVGNVANNVLDD430 pKa = 4.2GGAGSDD436 pKa = 3.36TVIFTNTTGINVTLNDD452 pKa = 3.23TGADD456 pKa = 3.31VVVSHH461 pKa = 7.45DD462 pKa = 4.12GEE464 pKa = 4.64TDD466 pKa = 3.32TLRR469 pKa = 11.84SIEE472 pKa = 4.38NIVGTSASDD481 pKa = 3.74TITGNSQDD489 pKa = 3.14NSLYY493 pKa = 10.75GGTGGADD500 pKa = 3.45TLSGGAGNDD509 pKa = 3.12RR510 pKa = 11.84LFGGGFTTTTTYY522 pKa = 10.73SAPSQADD529 pKa = 2.99ITKK532 pKa = 10.33AQSTNNGSIATAVATAGAFDD552 pKa = 3.98VDD554 pKa = 4.26ANPNIINSTSVPHH567 pKa = 5.71ATINATAAGGAVEE580 pKa = 4.29YY581 pKa = 10.85YY582 pKa = 10.35KK583 pKa = 10.42IEE585 pKa = 4.07VTQAGAQAVFDD596 pKa = 3.85MDD598 pKa = 4.11GNGTIGDD605 pKa = 4.16NIIEE609 pKa = 4.52LVNSTGTVLASNDD622 pKa = 4.12TGPGDD627 pKa = 4.6PGTTTNDD634 pKa = 3.45DD635 pKa = 3.44AYY637 pKa = 9.98IAYY640 pKa = 8.3TFATPGTYY648 pKa = 10.06YY649 pKa = 10.1IRR651 pKa = 11.84VGRR654 pKa = 11.84WISGSSAAQPLVAGQTYY671 pKa = 9.1QLHH674 pKa = 6.7ISLQGAAAVTTTVSANNTSSAVLDD698 pKa = 4.05GGAGNDD704 pKa = 3.81LLVGTLADD712 pKa = 4.23DD713 pKa = 4.56LLNGGSDD720 pKa = 3.6NDD722 pKa = 3.89TASFVNAFSGGSTTGVTVDD741 pKa = 4.69LNVQGVAQNTVAAGSDD757 pKa = 3.45TLSGIEE763 pKa = 3.97NLVGSGLNDD772 pKa = 3.57TLTGDD777 pKa = 3.66GNDD780 pKa = 3.53NVIEE784 pKa = 4.57GGLGNDD790 pKa = 3.64VLAGGAGVDD799 pKa = 3.51TASYY803 pKa = 10.18AGAAAGVAVSLALQGSAQNTVNAGTDD829 pKa = 3.43TLSGFEE835 pKa = 4.34NLSGSAFADD844 pKa = 3.74SLTGDD849 pKa = 3.41ASANVVSGGAGDD861 pKa = 4.05DD862 pKa = 3.84TLNPGANAAGAVDD875 pKa = 4.96LLDD878 pKa = 4.47GGAGADD884 pKa = 3.43TASFAGLGAVTVALNGAGDD903 pKa = 3.74ATATIGGAAVATLRR917 pKa = 11.84SIEE920 pKa = 4.01NLLGGANADD929 pKa = 4.25TLTGDD934 pKa = 3.56AGSNVIDD941 pKa = 4.57GGLGDD946 pKa = 5.2DD947 pKa = 4.15VLEE950 pKa = 4.92GGAGVDD956 pKa = 3.83TISFASAGSAAVTVNLATTTAQATGRR982 pKa = 11.84GSKK985 pKa = 9.37TISGFEE991 pKa = 3.92NVLTGTGNDD1000 pKa = 3.43AVTGDD1005 pKa = 3.53GGDD1008 pKa = 3.15NVFFEE1013 pKa = 4.63GRR1015 pKa = 11.84GNDD1018 pKa = 3.51VYY1020 pKa = 11.43NGAGGSDD1027 pKa = 3.36TLDD1030 pKa = 3.51YY1031 pKa = 10.91SASTSTVAMNLALATAQNTGTWGGTDD1057 pKa = 3.43TISNIEE1063 pKa = 4.19NFVGAATFANTVIGSAAANRR1083 pKa = 11.84LVGGSAVDD1091 pKa = 3.44TMEE1094 pKa = 4.19GRR1096 pKa = 11.84NGADD1100 pKa = 3.23VLIGGAGNDD1109 pKa = 3.71TLLGGTTGALDD1120 pKa = 4.42DD1121 pKa = 4.69GSADD1125 pKa = 3.65VLEE1128 pKa = 5.25GGAGSDD1134 pKa = 4.07FLSGGQGNDD1143 pKa = 2.75ILRR1146 pKa = 11.84GGDD1149 pKa = 3.56GDD1151 pKa = 4.33DD1152 pKa = 3.81SLVGG1156 pKa = 3.55
MM1 pKa = 7.38GGSRR5 pKa = 11.84MSTEE9 pKa = 4.38LGFDD13 pKa = 3.34APASNLVPLEE23 pKa = 4.67EE24 pKa = 4.28IAHH27 pKa = 4.77TTFVEE32 pKa = 4.87GCGCAACQGFQDD44 pKa = 4.08AGTDD48 pKa = 3.94GVISKK53 pKa = 10.96GGFNEE58 pKa = 4.2ANPEE62 pKa = 4.04SGGIGVLLPLVTNPDD77 pKa = 3.23GSRR80 pKa = 11.84SITGEE85 pKa = 3.97RR86 pKa = 11.84NVDD89 pKa = 3.97AILIGAKK96 pKa = 9.12WGATNLTFSFPKK108 pKa = 10.42SGSNYY113 pKa = 9.86NGTGFDD119 pKa = 3.86SNGVSIYY126 pKa = 10.77HH127 pKa = 6.11MEE129 pKa = 5.19LGAQQQAAARR139 pKa = 11.84AAFAQISAATGLTFTEE155 pKa = 4.14ITEE158 pKa = 4.3TDD160 pKa = 3.41TVHH163 pKa = 6.72ANIRR167 pKa = 11.84ISQSGDD173 pKa = 2.96NDD175 pKa = 3.36VTSAYY180 pKa = 10.54GGFPSDD186 pKa = 3.34TRR188 pKa = 11.84GVAGDD193 pKa = 2.84IWFGRR198 pKa = 11.84TSQPYY203 pKa = 9.8YY204 pKa = 10.86DD205 pKa = 4.37LAYY208 pKa = 10.03KK209 pKa = 7.78GTWGYY214 pKa = 8.84ATMMHH219 pKa = 7.03EE220 pKa = 4.66IGHH223 pKa = 5.19TMGLKK228 pKa = 10.05HH229 pKa = 6.31GHH231 pKa = 5.58QDD233 pKa = 3.44YY234 pKa = 9.86TNSDD238 pKa = 3.12LSFYY242 pKa = 10.68FGTSPRR248 pKa = 11.84RR249 pKa = 11.84GTQSLTPDD257 pKa = 3.43NDD259 pKa = 3.47GQAWSLMTYY268 pKa = 9.37TPAPFTNSNFAGEE281 pKa = 4.87KK282 pKa = 9.42VNQPQTYY289 pKa = 7.5MQYY292 pKa = 11.57DD293 pKa = 4.48LAALQYY299 pKa = 10.17MYY301 pKa = 10.94GANYY305 pKa = 7.39NTNAGDD311 pKa = 3.89TVYY314 pKa = 9.39TWSEE318 pKa = 3.99TTGEE322 pKa = 4.12MFVNGVGQGAPSGNTILLTIWDD344 pKa = 4.15GGGVDD349 pKa = 4.76TLDD352 pKa = 3.56LSNYY356 pKa = 9.71ADD358 pKa = 3.77GVTVDD363 pKa = 4.27LRR365 pKa = 11.84PGQFSTFDD373 pKa = 3.24QDD375 pKa = 3.52QLANHH380 pKa = 7.39LAYY383 pKa = 10.65QNLTALAPGNVAMSLLYY400 pKa = 11.17NNDD403 pKa = 2.66GRR405 pKa = 11.84SLIEE409 pKa = 3.83NAKK412 pKa = 10.49GGIGNDD418 pKa = 3.17VFVGNVANNVLDD430 pKa = 4.2GGAGSDD436 pKa = 3.36TVIFTNTTGINVTLNDD452 pKa = 3.23TGADD456 pKa = 3.31VVVSHH461 pKa = 7.45DD462 pKa = 4.12GEE464 pKa = 4.64TDD466 pKa = 3.32TLRR469 pKa = 11.84SIEE472 pKa = 4.38NIVGTSASDD481 pKa = 3.74TITGNSQDD489 pKa = 3.14NSLYY493 pKa = 10.75GGTGGADD500 pKa = 3.45TLSGGAGNDD509 pKa = 3.12RR510 pKa = 11.84LFGGGFTTTTTYY522 pKa = 10.73SAPSQADD529 pKa = 2.99ITKK532 pKa = 10.33AQSTNNGSIATAVATAGAFDD552 pKa = 3.98VDD554 pKa = 4.26ANPNIINSTSVPHH567 pKa = 5.71ATINATAAGGAVEE580 pKa = 4.29YY581 pKa = 10.85YY582 pKa = 10.35KK583 pKa = 10.42IEE585 pKa = 4.07VTQAGAQAVFDD596 pKa = 3.85MDD598 pKa = 4.11GNGTIGDD605 pKa = 4.16NIIEE609 pKa = 4.52LVNSTGTVLASNDD622 pKa = 4.12TGPGDD627 pKa = 4.6PGTTTNDD634 pKa = 3.45DD635 pKa = 3.44AYY637 pKa = 9.98IAYY640 pKa = 8.3TFATPGTYY648 pKa = 10.06YY649 pKa = 10.1IRR651 pKa = 11.84VGRR654 pKa = 11.84WISGSSAAQPLVAGQTYY671 pKa = 9.1QLHH674 pKa = 6.7ISLQGAAAVTTTVSANNTSSAVLDD698 pKa = 4.05GGAGNDD704 pKa = 3.81LLVGTLADD712 pKa = 4.23DD713 pKa = 4.56LLNGGSDD720 pKa = 3.6NDD722 pKa = 3.89TASFVNAFSGGSTTGVTVDD741 pKa = 4.69LNVQGVAQNTVAAGSDD757 pKa = 3.45TLSGIEE763 pKa = 3.97NLVGSGLNDD772 pKa = 3.57TLTGDD777 pKa = 3.66GNDD780 pKa = 3.53NVIEE784 pKa = 4.57GGLGNDD790 pKa = 3.64VLAGGAGVDD799 pKa = 3.51TASYY803 pKa = 10.18AGAAAGVAVSLALQGSAQNTVNAGTDD829 pKa = 3.43TLSGFEE835 pKa = 4.34NLSGSAFADD844 pKa = 3.74SLTGDD849 pKa = 3.41ASANVVSGGAGDD861 pKa = 4.05DD862 pKa = 3.84TLNPGANAAGAVDD875 pKa = 4.96LLDD878 pKa = 4.47GGAGADD884 pKa = 3.43TASFAGLGAVTVALNGAGDD903 pKa = 3.74ATATIGGAAVATLRR917 pKa = 11.84SIEE920 pKa = 4.01NLLGGANADD929 pKa = 4.25TLTGDD934 pKa = 3.56AGSNVIDD941 pKa = 4.57GGLGDD946 pKa = 5.2DD947 pKa = 4.15VLEE950 pKa = 4.92GGAGVDD956 pKa = 3.83TISFASAGSAAVTVNLATTTAQATGRR982 pKa = 11.84GSKK985 pKa = 9.37TISGFEE991 pKa = 3.92NVLTGTGNDD1000 pKa = 3.43AVTGDD1005 pKa = 3.53GGDD1008 pKa = 3.15NVFFEE1013 pKa = 4.63GRR1015 pKa = 11.84GNDD1018 pKa = 3.51VYY1020 pKa = 11.43NGAGGSDD1027 pKa = 3.36TLDD1030 pKa = 3.51YY1031 pKa = 10.91SASTSTVAMNLALATAQNTGTWGGTDD1057 pKa = 3.43TISNIEE1063 pKa = 4.19NFVGAATFANTVIGSAAANRR1083 pKa = 11.84LVGGSAVDD1091 pKa = 3.44TMEE1094 pKa = 4.19GRR1096 pKa = 11.84NGADD1100 pKa = 3.23VLIGGAGNDD1109 pKa = 3.71TLLGGTTGALDD1120 pKa = 4.42DD1121 pKa = 4.69GSADD1125 pKa = 3.65VLEE1128 pKa = 5.25GGAGSDD1134 pKa = 4.07FLSGGQGNDD1143 pKa = 2.75ILRR1146 pKa = 11.84GGDD1149 pKa = 3.56GDD1151 pKa = 4.33DD1152 pKa = 3.81SLVGG1156 pKa = 3.55
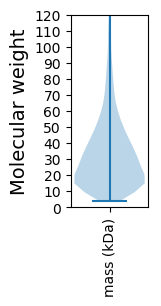
Molecular weight: 115.4 kDa
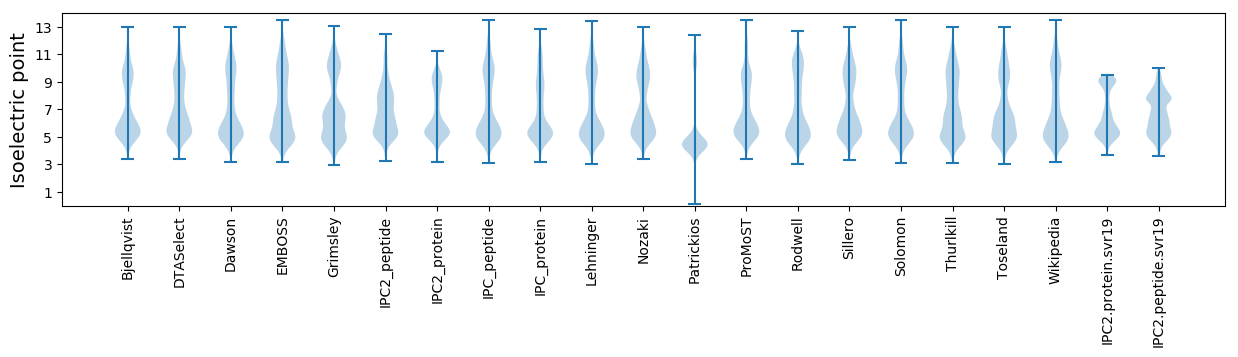
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y8ZPS1|A0A4Y8ZPS1_9SPHN UvrABC system protein B OS=Sphingomonas parva OX=2555898 GN=uvrB PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.51GFRR19 pKa = 11.84ARR21 pKa = 11.84KK22 pKa = 7.81ATVGGRR28 pKa = 11.84KK29 pKa = 9.05VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 6.76TLSAA44 pKa = 4.22
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.51GFRR19 pKa = 11.84ARR21 pKa = 11.84KK22 pKa = 7.81ATVGGRR28 pKa = 11.84KK29 pKa = 9.05VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 6.76TLSAA44 pKa = 4.22
Molecular weight: 5.05 kDa
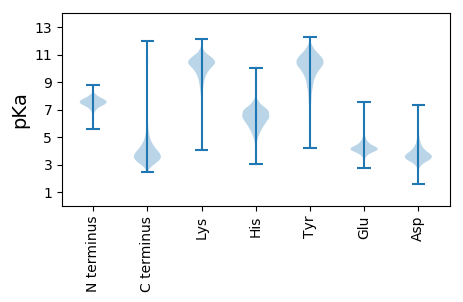
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1299525 |
34 |
5419 |
323.2 |
34.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.873 ± 0.064 | 0.758 ± 0.013 |
5.774 ± 0.049 | 5.82 ± 0.039 |
3.478 ± 0.022 | 9.22 ± 0.056 |
1.864 ± 0.019 | 4.651 ± 0.027 |
2.449 ± 0.027 | 10.145 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.078 ± 0.02 | 2.372 ± 0.033 |
5.475 ± 0.036 | 2.928 ± 0.024 |
8.162 ± 0.052 | 5.273 ± 0.034 |
5.024 ± 0.04 | 7.141 ± 0.034 |
1.431 ± 0.017 | 2.086 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
