
Pelagibacter phage HTVC022P
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; Pelagivirus; unclassified Pelagivirus
Average proteome isoelectric point is 7.06
Get precalculated fractions of proteins
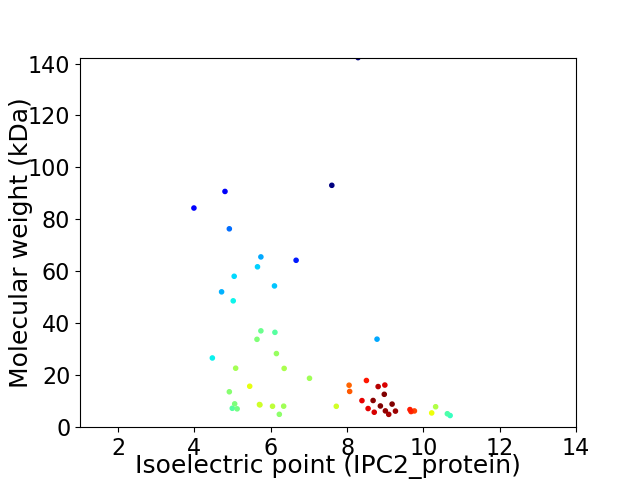
Virtual 2D-PAGE plot for 54 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y1NU94|A0A4Y1NU94_9CAUD Uncharacterized protein OS=Pelagibacter phage HTVC022P OX=2283016 GN=P022_gp26 PE=4 SV=1
MM1 pKa = 7.47ANSFVRR7 pKa = 11.84YY8 pKa = 8.02TGDD11 pKa = 3.14NSTTSYY17 pKa = 10.85SIPFSYY23 pKa = 10.6RR24 pKa = 11.84STADD28 pKa = 3.21LTVTLAGSVTTAFSLNSAGTTLTFNSAPAQDD59 pKa = 3.49AAIEE63 pKa = 4.02IRR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84TSQTTKK73 pKa = 10.65LVDD76 pKa = 3.6YY77 pKa = 10.81ASGSVLTEE85 pKa = 3.68NDD87 pKa = 3.94LDD89 pKa = 4.06TDD91 pKa = 3.63SDD93 pKa = 3.8QAFFMSQEE101 pKa = 4.56AIDD104 pKa = 4.32DD105 pKa = 4.22AGDD108 pKa = 3.66VIKK111 pKa = 11.1VSNTDD116 pKa = 4.06FQWDD120 pKa = 3.73AQNKK124 pKa = 9.11RR125 pKa = 11.84LTNVADD131 pKa = 3.69PTAAQHH137 pKa = 6.77AATKK141 pKa = 10.58NYY143 pKa = 10.6LEE145 pKa = 4.28NTWLSATDD153 pKa = 3.51KK154 pKa = 10.82ATLNTVNSNMTAINTVNSNITAIGTANTNSANITTVANNIGSVNTVATDD203 pKa = 3.42ISKK206 pKa = 10.95VIAVANDD213 pKa = 3.2LAEE216 pKa = 4.23AVSEE220 pKa = 4.31IEE222 pKa = 4.66TVADD226 pKa = 4.25DD227 pKa = 4.6LNEE230 pKa = 4.22STSEE234 pKa = 3.73IDD236 pKa = 3.59VVSNNIANVNIVGGISSDD254 pKa = 3.06ITSVAGIASAITAVNNNSTNINAVNTNSSNINTVAGDD291 pKa = 3.62STEE294 pKa = 4.08INAVAGNASNINSVAGITSDD314 pKa = 2.97ITSVAGISTAVTAVNNNSTNINAVNSNSSNINTVAGMQSAINTVNSSATAINAVNANATNINTVAGANTNITNVAGGLTNINTVATNLASVNNFAEE410 pKa = 4.49QYY412 pKa = 9.19RR413 pKa = 11.84ISSSAPTTSLNVGDD427 pKa = 5.41LYY429 pKa = 11.47FDD431 pKa = 3.51TTANEE436 pKa = 3.92LKK438 pKa = 10.5VYY440 pKa = 10.44KK441 pKa = 10.46SSGWAAAGSTVNGTSQRR458 pKa = 11.84YY459 pKa = 6.05TYY461 pKa = 10.25NITSAVSSVTGSDD474 pKa = 3.38ANGNTLAYY482 pKa = 9.86DD483 pKa = 3.68AGYY486 pKa = 11.18ADD488 pKa = 4.22VYY490 pKa = 11.21LNGVRR495 pKa = 11.84LSSSDD500 pKa = 3.02ITITSGTSVVFASALSNGDD519 pKa = 3.25VVDD522 pKa = 4.3IVAYY526 pKa = 8.81GTFNVASINAANIDD540 pKa = 3.67SGTINDD546 pKa = 3.73ARR548 pKa = 11.84LPTTMAGKK556 pKa = 8.26TLTTATVEE564 pKa = 4.34ANSLTARR571 pKa = 11.84GDD573 pKa = 3.45GSSADD578 pKa = 3.61GKK580 pKa = 9.41ITLNCSQNSHH590 pKa = 5.66GVSISSPNHH599 pKa = 6.46ASGQSYY605 pKa = 10.79NLILPTSVGTSGQVLATAGSNTNQLSWVDD634 pKa = 3.47ATEE637 pKa = 4.27TKK639 pKa = 9.17PTVADD644 pKa = 3.78VSQTIAPATATTINITGTNFVAIPIVEE671 pKa = 5.06FIKK674 pKa = 10.15TDD676 pKa = 3.55GSITLANTVSFTNATTLSVNVTLATGNYY704 pKa = 8.01HH705 pKa = 6.22VRR707 pKa = 11.84VEE709 pKa = 4.34NPDD712 pKa = 3.32GNAGRR717 pKa = 11.84STNNILTASTAPTFSTSAGSLGTIAGDD744 pKa = 3.67FSGTVATIAGSSDD757 pKa = 3.34SAVTFSEE764 pKa = 4.58TTSVLSTANCTLSSAGVITTTDD786 pKa = 3.7FGGSSTTATTYY797 pKa = 11.26NFIIRR802 pKa = 11.84ITDD805 pKa = 3.47AEE807 pKa = 4.45GQTADD812 pKa = 3.83RR813 pKa = 11.84SFSLTSSFGATGGGQFNN830 pKa = 4.04
MM1 pKa = 7.47ANSFVRR7 pKa = 11.84YY8 pKa = 8.02TGDD11 pKa = 3.14NSTTSYY17 pKa = 10.85SIPFSYY23 pKa = 10.6RR24 pKa = 11.84STADD28 pKa = 3.21LTVTLAGSVTTAFSLNSAGTTLTFNSAPAQDD59 pKa = 3.49AAIEE63 pKa = 4.02IRR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84TSQTTKK73 pKa = 10.65LVDD76 pKa = 3.6YY77 pKa = 10.81ASGSVLTEE85 pKa = 3.68NDD87 pKa = 3.94LDD89 pKa = 4.06TDD91 pKa = 3.63SDD93 pKa = 3.8QAFFMSQEE101 pKa = 4.56AIDD104 pKa = 4.32DD105 pKa = 4.22AGDD108 pKa = 3.66VIKK111 pKa = 11.1VSNTDD116 pKa = 4.06FQWDD120 pKa = 3.73AQNKK124 pKa = 9.11RR125 pKa = 11.84LTNVADD131 pKa = 3.69PTAAQHH137 pKa = 6.77AATKK141 pKa = 10.58NYY143 pKa = 10.6LEE145 pKa = 4.28NTWLSATDD153 pKa = 3.51KK154 pKa = 10.82ATLNTVNSNMTAINTVNSNITAIGTANTNSANITTVANNIGSVNTVATDD203 pKa = 3.42ISKK206 pKa = 10.95VIAVANDD213 pKa = 3.2LAEE216 pKa = 4.23AVSEE220 pKa = 4.31IEE222 pKa = 4.66TVADD226 pKa = 4.25DD227 pKa = 4.6LNEE230 pKa = 4.22STSEE234 pKa = 3.73IDD236 pKa = 3.59VVSNNIANVNIVGGISSDD254 pKa = 3.06ITSVAGIASAITAVNNNSTNINAVNTNSSNINTVAGDD291 pKa = 3.62STEE294 pKa = 4.08INAVAGNASNINSVAGITSDD314 pKa = 2.97ITSVAGISTAVTAVNNNSTNINAVNSNSSNINTVAGMQSAINTVNSSATAINAVNANATNINTVAGANTNITNVAGGLTNINTVATNLASVNNFAEE410 pKa = 4.49QYY412 pKa = 9.19RR413 pKa = 11.84ISSSAPTTSLNVGDD427 pKa = 5.41LYY429 pKa = 11.47FDD431 pKa = 3.51TTANEE436 pKa = 3.92LKK438 pKa = 10.5VYY440 pKa = 10.44KK441 pKa = 10.46SSGWAAAGSTVNGTSQRR458 pKa = 11.84YY459 pKa = 6.05TYY461 pKa = 10.25NITSAVSSVTGSDD474 pKa = 3.38ANGNTLAYY482 pKa = 9.86DD483 pKa = 3.68AGYY486 pKa = 11.18ADD488 pKa = 4.22VYY490 pKa = 11.21LNGVRR495 pKa = 11.84LSSSDD500 pKa = 3.02ITITSGTSVVFASALSNGDD519 pKa = 3.25VVDD522 pKa = 4.3IVAYY526 pKa = 8.81GTFNVASINAANIDD540 pKa = 3.67SGTINDD546 pKa = 3.73ARR548 pKa = 11.84LPTTMAGKK556 pKa = 8.26TLTTATVEE564 pKa = 4.34ANSLTARR571 pKa = 11.84GDD573 pKa = 3.45GSSADD578 pKa = 3.61GKK580 pKa = 9.41ITLNCSQNSHH590 pKa = 5.66GVSISSPNHH599 pKa = 6.46ASGQSYY605 pKa = 10.79NLILPTSVGTSGQVLATAGSNTNQLSWVDD634 pKa = 3.47ATEE637 pKa = 4.27TKK639 pKa = 9.17PTVADD644 pKa = 3.78VSQTIAPATATTINITGTNFVAIPIVEE671 pKa = 5.06FIKK674 pKa = 10.15TDD676 pKa = 3.55GSITLANTVSFTNATTLSVNVTLATGNYY704 pKa = 8.01HH705 pKa = 6.22VRR707 pKa = 11.84VEE709 pKa = 4.34NPDD712 pKa = 3.32GNAGRR717 pKa = 11.84STNNILTASTAPTFSTSAGSLGTIAGDD744 pKa = 3.67FSGTVATIAGSSDD757 pKa = 3.34SAVTFSEE764 pKa = 4.58TTSVLSTANCTLSSAGVITTTDD786 pKa = 3.7FGGSSTTATTYY797 pKa = 11.26NFIIRR802 pKa = 11.84ITDD805 pKa = 3.47AEE807 pKa = 4.45GQTADD812 pKa = 3.83RR813 pKa = 11.84SFSLTSSFGATGGGQFNN830 pKa = 4.04
Molecular weight: 84.38 kDa
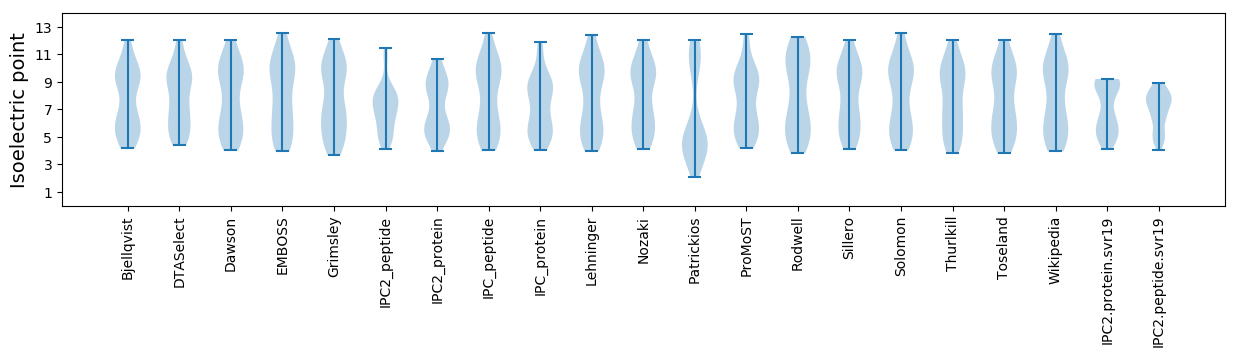
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y1NUD1|A0A4Y1NUD1_9CAUD Lysozyme murein hydrolase OS=Pelagibacter phage HTVC022P OX=2283016 GN=P022_gp54 PE=4 SV=1
MM1 pKa = 7.34ARR3 pKa = 11.84KK4 pKa = 9.32FKK6 pKa = 11.11DD7 pKa = 3.37FVVRR11 pKa = 11.84EE12 pKa = 4.09KK13 pKa = 10.49PKK15 pKa = 10.68KK16 pKa = 9.69RR17 pKa = 11.84VRR19 pKa = 11.84THH21 pKa = 7.01KK22 pKa = 10.52KK23 pKa = 9.88RR24 pKa = 11.84LNKK27 pKa = 9.92NEE29 pKa = 3.3KK30 pKa = 9.69RR31 pKa = 11.84DD32 pKa = 3.66YY33 pKa = 11.04KK34 pKa = 10.71KK35 pKa = 10.79YY36 pKa = 10.14NRR38 pKa = 11.84QGRR41 pKa = 11.84PQQ43 pKa = 3.09
MM1 pKa = 7.34ARR3 pKa = 11.84KK4 pKa = 9.32FKK6 pKa = 11.11DD7 pKa = 3.37FVVRR11 pKa = 11.84EE12 pKa = 4.09KK13 pKa = 10.49PKK15 pKa = 10.68KK16 pKa = 9.69RR17 pKa = 11.84VRR19 pKa = 11.84THH21 pKa = 7.01KK22 pKa = 10.52KK23 pKa = 9.88RR24 pKa = 11.84LNKK27 pKa = 9.92NEE29 pKa = 3.3KK30 pKa = 9.69RR31 pKa = 11.84DD32 pKa = 3.66YY33 pKa = 11.04KK34 pKa = 10.71KK35 pKa = 10.79YY36 pKa = 10.14NRR38 pKa = 11.84QGRR41 pKa = 11.84PQQ43 pKa = 3.09
Molecular weight: 5.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13037 |
40 |
1261 |
241.4 |
26.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.77 ± 0.397 | 0.79 ± 0.122 |
6.037 ± 0.231 | 6.236 ± 0.447 |
3.881 ± 0.21 | 6.689 ± 0.358 |
1.711 ± 0.163 | 6.359 ± 0.2 |
8.23 ± 0.607 | 7.74 ± 0.359 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.715 ± 0.272 | 6.152 ± 0.436 |
2.938 ± 0.179 | 3.682 ± 0.273 |
3.981 ± 0.296 | 7.348 ± 0.53 |
7.371 ± 0.653 | 5.738 ± 0.294 |
1.158 ± 0.142 | 3.475 ± 0.22 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
