
Hubei sobemo-like virus 34
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.52
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
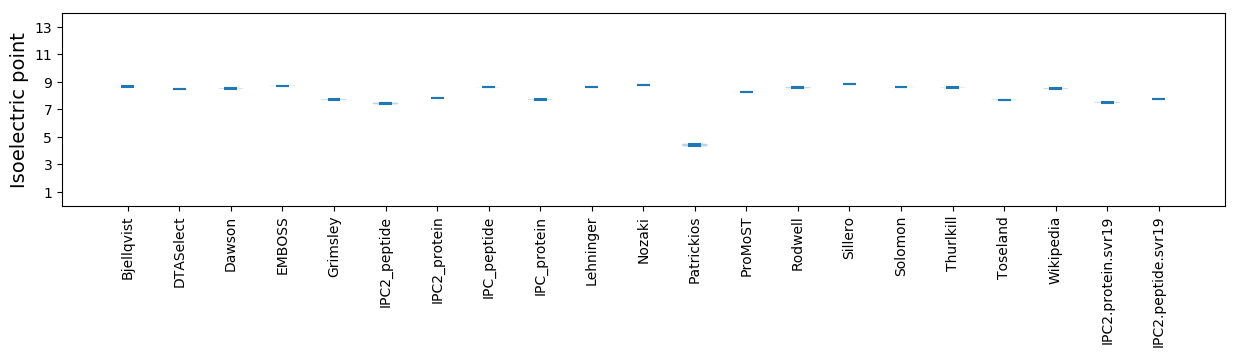
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KEP2|A0A1L3KEP2_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 34 OX=1923221 PE=4 SV=1
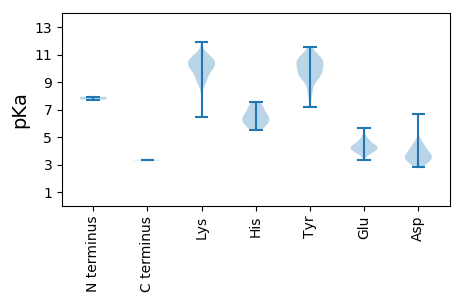
MM1 pKa = 7.93PGTSNVEE8 pKa = 4.21SIMDD12 pKa = 3.56IVILKK17 pKa = 9.71QLVIIGKK24 pKa = 8.9CLLVVGIAAGTVFMSVGLYY43 pKa = 9.7TLFSLIPGTIKK54 pKa = 10.82NMLVNWPRR62 pKa = 11.84EE63 pKa = 4.06MVQAKK68 pKa = 9.18YY69 pKa = 10.29RR70 pKa = 11.84EE71 pKa = 4.02YY72 pKa = 10.8LEE74 pKa = 4.9PYY76 pKa = 9.96GLVEE80 pKa = 4.58EE81 pKa = 4.8KK82 pKa = 10.32PEE84 pKa = 3.89PVIPKK89 pKa = 8.98PIKK92 pKa = 9.09IEE94 pKa = 3.77PVRR97 pKa = 11.84VEE99 pKa = 4.34IEE101 pKa = 4.21SLLQRR106 pKa = 11.84CYY108 pKa = 10.73IEE110 pKa = 4.3VCEE113 pKa = 4.9SIYY116 pKa = 11.39LMIGWIFINKK126 pKa = 6.46YY127 pKa = 7.19TVILMCILSLIILVIYY143 pKa = 10.2RR144 pKa = 11.84RR145 pKa = 11.84YY146 pKa = 9.58FRR148 pKa = 11.84HH149 pKa = 6.0FVSKK153 pKa = 10.52YY154 pKa = 8.21YY155 pKa = 10.84KK156 pKa = 9.22KK157 pKa = 8.89TVWWMRR163 pKa = 11.84GVDD166 pKa = 3.95TVTPPYY172 pKa = 9.52RR173 pKa = 11.84VEE175 pKa = 4.32SIIPGSDD182 pKa = 3.48FNPDD186 pKa = 3.18MVIPSYY192 pKa = 10.03QVEE195 pKa = 4.2IHH197 pKa = 6.97SSGIWTDD204 pKa = 2.83KK205 pKa = 10.08FVAYY209 pKa = 9.68GVRR212 pKa = 11.84FHH214 pKa = 7.55DD215 pKa = 4.08FLVTPRR221 pKa = 11.84HH222 pKa = 5.49VLTAVSNLKK231 pKa = 10.54GEE233 pKa = 4.41LMIKK237 pKa = 7.53TRR239 pKa = 11.84RR240 pKa = 11.84GSKK243 pKa = 9.94IIRR246 pKa = 11.84SQIMDD251 pKa = 4.15SIGLNDD257 pKa = 3.37MAYY260 pKa = 9.55IKK262 pKa = 10.7LEE264 pKa = 3.95SSIWADD270 pKa = 3.5LGVTSPSFPKK280 pKa = 10.44QLTEE284 pKa = 4.15VPSSVVTCVGKK295 pKa = 9.74PGGSTGSLKK304 pKa = 9.93FTDD307 pKa = 6.65FMGFWNYY314 pKa = 9.53TGSTVPGMSGAAYY327 pKa = 9.87FIGNVCYY334 pKa = 9.95GVHH337 pKa = 5.87QGVIGNINAGAMSLAIVEE355 pKa = 4.33EE356 pKa = 4.41LKK358 pKa = 11.03VLYY361 pKa = 10.62NPEE364 pKa = 4.37GYY366 pKa = 10.81KK367 pKa = 9.28MPRR370 pKa = 11.84SSTTPLNSDD379 pKa = 2.95AKK381 pKa = 8.47EE382 pKa = 4.47TEE384 pKa = 3.96FEE386 pKa = 4.62TPFTRR391 pKa = 11.84PKK393 pKa = 10.6KK394 pKa = 8.72YY395 pKa = 9.13TKK397 pKa = 10.05EE398 pKa = 3.97DD399 pKa = 3.35YY400 pKa = 11.56AEE402 pKa = 4.52FIKK405 pKa = 10.62KK406 pKa = 9.33KK407 pKa = 9.08QAEE410 pKa = 4.16GDD412 pKa = 3.2LWADD416 pKa = 3.8DD417 pKa = 3.91KK418 pKa = 11.95PEE420 pKa = 4.11FEE422 pKa = 4.74SNRR425 pKa = 11.84KK426 pKa = 9.69GEE428 pKa = 4.78AKK430 pKa = 10.34PEE432 pKa = 3.94MTQEE436 pKa = 3.82EE437 pKa = 5.66LFFKK441 pKa = 10.65ILGLTPEE448 pKa = 4.34QARR451 pKa = 11.84KK452 pKa = 9.39LLPFIDD458 pKa = 3.71QLARR462 pKa = 11.84TPTSTQQPPSEE473 pKa = 4.17EE474 pKa = 4.21KK475 pKa = 8.27MTGHH479 pKa = 6.35SPDD482 pKa = 3.73GPQIPVTKK490 pKa = 9.41IDD492 pKa = 3.32LEE494 pKa = 4.44TRR496 pKa = 11.84VSILEE501 pKa = 3.86NSKK504 pKa = 10.07VRR506 pKa = 11.84LEE508 pKa = 4.5AEE510 pKa = 3.76NSVLKK515 pKa = 10.77EE516 pKa = 4.02KK517 pKa = 10.8VKK519 pKa = 10.69HH520 pKa = 6.28LEE522 pKa = 3.81DD523 pKa = 5.58HH524 pKa = 6.6IDD526 pKa = 4.29KK527 pKa = 10.37IARR530 pKa = 11.84WSVLHH535 pKa = 6.05YY536 pKa = 10.72GFGRR540 pKa = 11.84KK541 pKa = 9.13KK542 pKa = 10.34YY543 pKa = 9.33EE544 pKa = 4.17CAFPQCDD551 pKa = 3.39RR552 pKa = 11.84IFVGKK557 pKa = 9.52QALIAHH563 pKa = 7.3KK564 pKa = 10.11LASDD568 pKa = 3.71HH569 pKa = 6.8LLEE572 pKa = 5.22GDD574 pKa = 3.85KK575 pKa = 10.96LLPNPVVQGEE585 pKa = 4.63SAFPADD591 pKa = 3.52NKK593 pKa = 10.17TKK595 pKa = 10.92EE596 pKa = 3.93EE597 pKa = 4.41LGFQKK602 pKa = 10.7KK603 pKa = 9.12PLQKK607 pKa = 10.2NHH609 pKa = 5.71QPKK612 pKa = 10.31QSGTPLRR619 pKa = 11.84NTLNMPEE626 pKa = 4.21KK627 pKa = 9.6PKK629 pKa = 10.58PSTSTQEE636 pKa = 4.17DD637 pKa = 4.2PNDD640 pKa = 3.85LKK642 pKa = 10.68TMMKK646 pKa = 9.85EE647 pKa = 3.46MHH649 pKa = 7.22KK650 pKa = 10.1ILQILVQKK658 pKa = 10.82SVGPNSATTQNN669 pKa = 3.34
MM1 pKa = 7.93PGTSNVEE8 pKa = 4.21SIMDD12 pKa = 3.56IVILKK17 pKa = 9.71QLVIIGKK24 pKa = 8.9CLLVVGIAAGTVFMSVGLYY43 pKa = 9.7TLFSLIPGTIKK54 pKa = 10.82NMLVNWPRR62 pKa = 11.84EE63 pKa = 4.06MVQAKK68 pKa = 9.18YY69 pKa = 10.29RR70 pKa = 11.84EE71 pKa = 4.02YY72 pKa = 10.8LEE74 pKa = 4.9PYY76 pKa = 9.96GLVEE80 pKa = 4.58EE81 pKa = 4.8KK82 pKa = 10.32PEE84 pKa = 3.89PVIPKK89 pKa = 8.98PIKK92 pKa = 9.09IEE94 pKa = 3.77PVRR97 pKa = 11.84VEE99 pKa = 4.34IEE101 pKa = 4.21SLLQRR106 pKa = 11.84CYY108 pKa = 10.73IEE110 pKa = 4.3VCEE113 pKa = 4.9SIYY116 pKa = 11.39LMIGWIFINKK126 pKa = 6.46YY127 pKa = 7.19TVILMCILSLIILVIYY143 pKa = 10.2RR144 pKa = 11.84RR145 pKa = 11.84YY146 pKa = 9.58FRR148 pKa = 11.84HH149 pKa = 6.0FVSKK153 pKa = 10.52YY154 pKa = 8.21YY155 pKa = 10.84KK156 pKa = 9.22KK157 pKa = 8.89TVWWMRR163 pKa = 11.84GVDD166 pKa = 3.95TVTPPYY172 pKa = 9.52RR173 pKa = 11.84VEE175 pKa = 4.32SIIPGSDD182 pKa = 3.48FNPDD186 pKa = 3.18MVIPSYY192 pKa = 10.03QVEE195 pKa = 4.2IHH197 pKa = 6.97SSGIWTDD204 pKa = 2.83KK205 pKa = 10.08FVAYY209 pKa = 9.68GVRR212 pKa = 11.84FHH214 pKa = 7.55DD215 pKa = 4.08FLVTPRR221 pKa = 11.84HH222 pKa = 5.49VLTAVSNLKK231 pKa = 10.54GEE233 pKa = 4.41LMIKK237 pKa = 7.53TRR239 pKa = 11.84RR240 pKa = 11.84GSKK243 pKa = 9.94IIRR246 pKa = 11.84SQIMDD251 pKa = 4.15SIGLNDD257 pKa = 3.37MAYY260 pKa = 9.55IKK262 pKa = 10.7LEE264 pKa = 3.95SSIWADD270 pKa = 3.5LGVTSPSFPKK280 pKa = 10.44QLTEE284 pKa = 4.15VPSSVVTCVGKK295 pKa = 9.74PGGSTGSLKK304 pKa = 9.93FTDD307 pKa = 6.65FMGFWNYY314 pKa = 9.53TGSTVPGMSGAAYY327 pKa = 9.87FIGNVCYY334 pKa = 9.95GVHH337 pKa = 5.87QGVIGNINAGAMSLAIVEE355 pKa = 4.33EE356 pKa = 4.41LKK358 pKa = 11.03VLYY361 pKa = 10.62NPEE364 pKa = 4.37GYY366 pKa = 10.81KK367 pKa = 9.28MPRR370 pKa = 11.84SSTTPLNSDD379 pKa = 2.95AKK381 pKa = 8.47EE382 pKa = 4.47TEE384 pKa = 3.96FEE386 pKa = 4.62TPFTRR391 pKa = 11.84PKK393 pKa = 10.6KK394 pKa = 8.72YY395 pKa = 9.13TKK397 pKa = 10.05EE398 pKa = 3.97DD399 pKa = 3.35YY400 pKa = 11.56AEE402 pKa = 4.52FIKK405 pKa = 10.62KK406 pKa = 9.33KK407 pKa = 9.08QAEE410 pKa = 4.16GDD412 pKa = 3.2LWADD416 pKa = 3.8DD417 pKa = 3.91KK418 pKa = 11.95PEE420 pKa = 4.11FEE422 pKa = 4.74SNRR425 pKa = 11.84KK426 pKa = 9.69GEE428 pKa = 4.78AKK430 pKa = 10.34PEE432 pKa = 3.94MTQEE436 pKa = 3.82EE437 pKa = 5.66LFFKK441 pKa = 10.65ILGLTPEE448 pKa = 4.34QARR451 pKa = 11.84KK452 pKa = 9.39LLPFIDD458 pKa = 3.71QLARR462 pKa = 11.84TPTSTQQPPSEE473 pKa = 4.17EE474 pKa = 4.21KK475 pKa = 8.27MTGHH479 pKa = 6.35SPDD482 pKa = 3.73GPQIPVTKK490 pKa = 9.41IDD492 pKa = 3.32LEE494 pKa = 4.44TRR496 pKa = 11.84VSILEE501 pKa = 3.86NSKK504 pKa = 10.07VRR506 pKa = 11.84LEE508 pKa = 4.5AEE510 pKa = 3.76NSVLKK515 pKa = 10.77EE516 pKa = 4.02KK517 pKa = 10.8VKK519 pKa = 10.69HH520 pKa = 6.28LEE522 pKa = 3.81DD523 pKa = 5.58HH524 pKa = 6.6IDD526 pKa = 4.29KK527 pKa = 10.37IARR530 pKa = 11.84WSVLHH535 pKa = 6.05YY536 pKa = 10.72GFGRR540 pKa = 11.84KK541 pKa = 9.13KK542 pKa = 10.34YY543 pKa = 9.33EE544 pKa = 4.17CAFPQCDD551 pKa = 3.39RR552 pKa = 11.84IFVGKK557 pKa = 9.52QALIAHH563 pKa = 7.3KK564 pKa = 10.11LASDD568 pKa = 3.71HH569 pKa = 6.8LLEE572 pKa = 5.22GDD574 pKa = 3.85KK575 pKa = 10.96LLPNPVVQGEE585 pKa = 4.63SAFPADD591 pKa = 3.52NKK593 pKa = 10.17TKK595 pKa = 10.92EE596 pKa = 3.93EE597 pKa = 4.41LGFQKK602 pKa = 10.7KK603 pKa = 9.12PLQKK607 pKa = 10.2NHH609 pKa = 5.71QPKK612 pKa = 10.31QSGTPLRR619 pKa = 11.84NTLNMPEE626 pKa = 4.21KK627 pKa = 9.6PKK629 pKa = 10.58PSTSTQEE636 pKa = 4.17DD637 pKa = 4.2PNDD640 pKa = 3.85LKK642 pKa = 10.68TMMKK646 pKa = 9.85EE647 pKa = 3.46MHH649 pKa = 7.22KK650 pKa = 10.1ILQILVQKK658 pKa = 10.82SVGPNSATTQNN669 pKa = 3.34
Molecular weight: 75.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEP2|A0A1L3KEP2_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 34 OX=1923221 PE=4 SV=1
MM1 pKa = 7.73SYY3 pKa = 9.39NHH5 pKa = 5.82YY6 pKa = 10.31VRR8 pKa = 11.84VVSEE12 pKa = 4.63LEE14 pKa = 4.29WNSSPGVPYY23 pKa = 10.48CHH25 pKa = 6.44QFTTNRR31 pKa = 11.84ILFGVKK37 pKa = 9.84DD38 pKa = 3.94GQPSQEE44 pKa = 3.89RR45 pKa = 11.84LKK47 pKa = 10.13MVWEE51 pKa = 4.33IVSKK55 pKa = 10.75RR56 pKa = 11.84LQDD59 pKa = 3.45QDD61 pKa = 3.42SDD63 pKa = 4.56PIRR66 pKa = 11.84LFVKK70 pKa = 10.11PEE72 pKa = 3.61PHH74 pKa = 6.73KK75 pKa = 10.73KK76 pKa = 10.46SKK78 pKa = 10.16IDD80 pKa = 3.33NKK82 pKa = 10.49AYY84 pKa = 10.71RR85 pKa = 11.84LISSVSVVDD94 pKa = 4.52QIIDD98 pKa = 3.33HH99 pKa = 6.28MLFDD103 pKa = 4.64HH104 pKa = 6.41MNKK107 pKa = 9.86KK108 pKa = 10.36ALDD111 pKa = 3.69NWWNNPVKK119 pKa = 10.72VGWSPVVGGWKK130 pKa = 10.19FIPTNYY136 pKa = 9.73ISTDD140 pKa = 3.0KK141 pKa = 11.3SSWDD145 pKa = 3.15WTVGMWLIEE154 pKa = 4.02LVFEE158 pKa = 4.79LRR160 pKa = 11.84KK161 pKa = 10.17DD162 pKa = 3.44LCVTKK167 pKa = 10.85GPLYY171 pKa = 10.64DD172 pKa = 2.89KK173 pKa = 10.1WLRR176 pKa = 11.84LAIFRR181 pKa = 11.84FTKK184 pKa = 10.47LFYY187 pKa = 10.74DD188 pKa = 4.46FQLVTSGGYY197 pKa = 8.33KK198 pKa = 10.03LRR200 pKa = 11.84YY201 pKa = 8.33KK202 pKa = 10.73GHH204 pKa = 7.19GIMKK208 pKa = 9.41SGCVNTLVDD217 pKa = 3.81NSLMQLLLHH226 pKa = 6.11VMVSFRR232 pKa = 11.84LNKK235 pKa = 10.37SPVDD239 pKa = 3.38IMAMGDD245 pKa = 3.62DD246 pKa = 4.36VIMEE250 pKa = 4.97FIDD253 pKa = 3.69YY254 pKa = 10.68LEE256 pKa = 4.87EE257 pKa = 3.94YY258 pKa = 10.41LAEE261 pKa = 4.29TKK263 pKa = 10.0KK264 pKa = 10.13HH265 pKa = 6.04CIIKK269 pKa = 9.87EE270 pKa = 4.16VNFQTEE276 pKa = 4.35FAGLRR281 pKa = 11.84YY282 pKa = 9.22KK283 pKa = 10.85KK284 pKa = 10.5NGIIEE289 pKa = 3.93PLYY292 pKa = 9.07TGKK295 pKa = 9.7HH296 pKa = 5.56AYY298 pKa = 9.29QLLHH302 pKa = 6.42MPNKK306 pKa = 9.76FKK308 pKa = 11.56NEE310 pKa = 3.37IAASYY315 pKa = 10.14SLLYY319 pKa = 10.31HH320 pKa = 7.16RR321 pKa = 11.84STHH324 pKa = 6.64RR325 pKa = 11.84DD326 pKa = 2.94WIRR329 pKa = 11.84QKK331 pKa = 11.08LEE333 pKa = 4.29SIGCNLPPLYY343 pKa = 10.41EE344 pKa = 5.33LDD346 pKa = 4.62AIFDD350 pKa = 4.3GEE352 pKa = 4.43CC353 pKa = 3.3
MM1 pKa = 7.73SYY3 pKa = 9.39NHH5 pKa = 5.82YY6 pKa = 10.31VRR8 pKa = 11.84VVSEE12 pKa = 4.63LEE14 pKa = 4.29WNSSPGVPYY23 pKa = 10.48CHH25 pKa = 6.44QFTTNRR31 pKa = 11.84ILFGVKK37 pKa = 9.84DD38 pKa = 3.94GQPSQEE44 pKa = 3.89RR45 pKa = 11.84LKK47 pKa = 10.13MVWEE51 pKa = 4.33IVSKK55 pKa = 10.75RR56 pKa = 11.84LQDD59 pKa = 3.45QDD61 pKa = 3.42SDD63 pKa = 4.56PIRR66 pKa = 11.84LFVKK70 pKa = 10.11PEE72 pKa = 3.61PHH74 pKa = 6.73KK75 pKa = 10.73KK76 pKa = 10.46SKK78 pKa = 10.16IDD80 pKa = 3.33NKK82 pKa = 10.49AYY84 pKa = 10.71RR85 pKa = 11.84LISSVSVVDD94 pKa = 4.52QIIDD98 pKa = 3.33HH99 pKa = 6.28MLFDD103 pKa = 4.64HH104 pKa = 6.41MNKK107 pKa = 9.86KK108 pKa = 10.36ALDD111 pKa = 3.69NWWNNPVKK119 pKa = 10.72VGWSPVVGGWKK130 pKa = 10.19FIPTNYY136 pKa = 9.73ISTDD140 pKa = 3.0KK141 pKa = 11.3SSWDD145 pKa = 3.15WTVGMWLIEE154 pKa = 4.02LVFEE158 pKa = 4.79LRR160 pKa = 11.84KK161 pKa = 10.17DD162 pKa = 3.44LCVTKK167 pKa = 10.85GPLYY171 pKa = 10.64DD172 pKa = 2.89KK173 pKa = 10.1WLRR176 pKa = 11.84LAIFRR181 pKa = 11.84FTKK184 pKa = 10.47LFYY187 pKa = 10.74DD188 pKa = 4.46FQLVTSGGYY197 pKa = 8.33KK198 pKa = 10.03LRR200 pKa = 11.84YY201 pKa = 8.33KK202 pKa = 10.73GHH204 pKa = 7.19GIMKK208 pKa = 9.41SGCVNTLVDD217 pKa = 3.81NSLMQLLLHH226 pKa = 6.11VMVSFRR232 pKa = 11.84LNKK235 pKa = 10.37SPVDD239 pKa = 3.38IMAMGDD245 pKa = 3.62DD246 pKa = 4.36VIMEE250 pKa = 4.97FIDD253 pKa = 3.69YY254 pKa = 10.68LEE256 pKa = 4.87EE257 pKa = 3.94YY258 pKa = 10.41LAEE261 pKa = 4.29TKK263 pKa = 10.0KK264 pKa = 10.13HH265 pKa = 6.04CIIKK269 pKa = 9.87EE270 pKa = 4.16VNFQTEE276 pKa = 4.35FAGLRR281 pKa = 11.84YY282 pKa = 9.22KK283 pKa = 10.85KK284 pKa = 10.5NGIIEE289 pKa = 3.93PLYY292 pKa = 9.07TGKK295 pKa = 9.7HH296 pKa = 5.56AYY298 pKa = 9.29QLLHH302 pKa = 6.42MPNKK306 pKa = 9.76FKK308 pKa = 11.56NEE310 pKa = 3.37IAASYY315 pKa = 10.14SLLYY319 pKa = 10.31HH320 pKa = 7.16RR321 pKa = 11.84STHH324 pKa = 6.64RR325 pKa = 11.84DD326 pKa = 2.94WIRR329 pKa = 11.84QKK331 pKa = 11.08LEE333 pKa = 4.29SIGCNLPPLYY343 pKa = 10.41EE344 pKa = 5.33LDD346 pKa = 4.62AIFDD350 pKa = 4.3GEE352 pKa = 4.43CC353 pKa = 3.3
Molecular weight: 41.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1022 |
353 |
669 |
511.0 |
58.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.718 ± 0.466 | 1.37 ± 0.174 |
4.697 ± 0.808 | 6.556 ± 0.767 |
4.11 ± 0.223 | 6.067 ± 0.36 |
2.446 ± 0.502 | 7.143 ± 0.33 |
8.513 ± 0.007 | 9.002 ± 0.63 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.327 ± 0.038 | 4.012 ± 0.423 |
6.067 ± 0.957 | 3.523 ± 0.214 |
3.914 ± 0.177 | 6.849 ± 0.027 |
5.284 ± 0.843 | 7.436 ± 0.037 |
1.957 ± 0.61 | 4.012 ± 0.423 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
