
Phytoactinopolyspora halotolerans
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Jiangellales; Jiangellaceae; Phytoactinopolyspora
Average proteome isoelectric point is 5.91
Get precalculated fractions of proteins
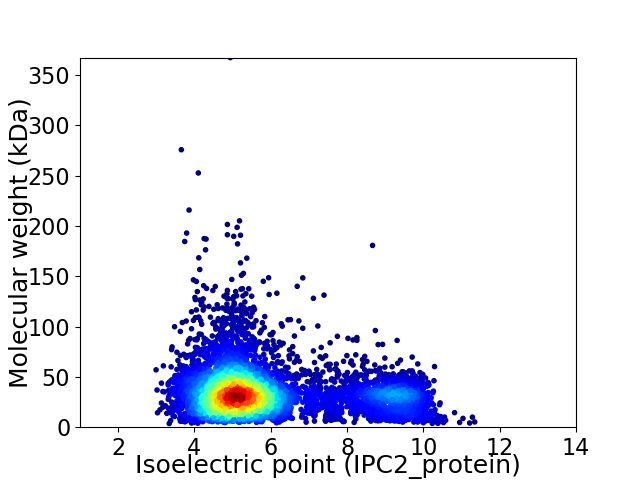
Virtual 2D-PAGE plot for 6265 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6L9SKN1|A0A6L9SKN1_9ACTN DoxX family protein OS=Phytoactinopolyspora halotolerans OX=1981512 GN=G1H10_31360 PE=4 SV=1
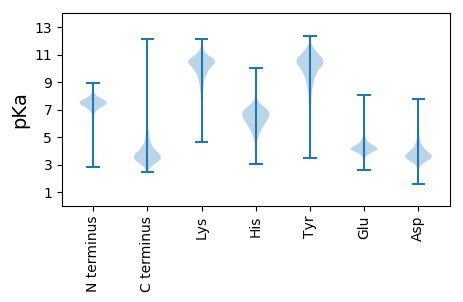
MM1 pKa = 7.73RR2 pKa = 11.84FDD4 pKa = 3.34RR5 pKa = 11.84HH6 pKa = 5.79SGRR9 pKa = 11.84HH10 pKa = 4.66RR11 pKa = 11.84LAGASALLAATGLMAGGLIATVATSAHH38 pKa = 6.12AAVACEE44 pKa = 3.86VDD46 pKa = 3.73YY47 pKa = 11.48SANTWTDD54 pKa = 3.35GASSGGFTASVTINNLGDD72 pKa = 3.9PVEE75 pKa = 4.45GWALEE80 pKa = 3.97FEE82 pKa = 4.89LPDD85 pKa = 3.9GQSFTQGWSANWDD98 pKa = 3.44ADD100 pKa = 3.73GTSITAEE107 pKa = 3.71NLAWNGTLGTGASTSVGFNGSWTGSYY133 pKa = 9.77TDD135 pKa = 4.34PSAFTLNGVACTGDD149 pKa = 3.67PGNGGDD155 pKa = 4.17NLPPTVSVTSPSHH168 pKa = 5.01GAEE171 pKa = 3.95YY172 pKa = 10.83DD173 pKa = 3.36EE174 pKa = 5.27GATVPLAATASDD186 pKa = 3.96PDD188 pKa = 3.74GTIDD192 pKa = 3.59RR193 pKa = 11.84VEE195 pKa = 4.05FLVDD199 pKa = 3.31GDD201 pKa = 4.36VVATDD206 pKa = 3.7ASAPYY211 pKa = 9.02EE212 pKa = 4.14ATVSGLDD219 pKa = 3.72TGSHH223 pKa = 4.98TASAVAYY230 pKa = 10.15DD231 pKa = 4.7DD232 pKa = 4.89GDD234 pKa = 4.32PEE236 pKa = 5.27LSATDD241 pKa = 3.56SASFTIGDD249 pKa = 4.6DD250 pKa = 4.1GEE252 pKa = 4.43EE253 pKa = 4.45PPPDD257 pKa = 3.8SDD259 pKa = 4.2NPYY262 pKa = 10.62AGADD266 pKa = 3.57VYY268 pKa = 11.68VNPDD272 pKa = 2.6WAGQVLDD279 pKa = 4.51EE280 pKa = 4.77ASSTGGALGAAMEE293 pKa = 4.49EE294 pKa = 4.69VATYY298 pKa = 7.49PTFVWMDD305 pKa = 4.32RR306 pKa = 11.84IGAITDD312 pKa = 3.33GTGLVGHH319 pKa = 7.63LDD321 pKa = 3.4NAVAQDD327 pKa = 3.71QANGATPTVFQVVIYY342 pKa = 9.25NLPNRR347 pKa = 11.84DD348 pKa = 3.57CAASASNGEE357 pKa = 4.29LLISEE362 pKa = 4.92NGFQRR367 pKa = 11.84YY368 pKa = 8.29RR369 pKa = 11.84EE370 pKa = 4.1EE371 pKa = 4.6YY372 pKa = 8.85IDD374 pKa = 5.06PIAEE378 pKa = 3.75ILSRR382 pKa = 11.84PEE384 pKa = 3.74YY385 pKa = 10.28SDD387 pKa = 4.89LRR389 pKa = 11.84IATVIEE395 pKa = 4.32VDD397 pKa = 3.7SLPNLVTNSDD407 pKa = 4.31DD408 pKa = 4.25PDD410 pKa = 3.86CQEE413 pKa = 3.79AAGDD417 pKa = 3.42GGYY420 pKa = 10.03RR421 pKa = 11.84DD422 pKa = 4.56GIRR425 pKa = 11.84YY426 pKa = 8.89ALNEE430 pKa = 3.89LSAIPNVYY438 pKa = 10.38NYY440 pKa = 10.97LDD442 pKa = 3.68IAHH445 pKa = 6.58SGWLGWDD452 pKa = 3.66SNFGPAVDD460 pKa = 5.74LYY462 pKa = 10.65VQTLTSSDD470 pKa = 3.9FYY472 pKa = 11.69GPAPGFDD479 pKa = 3.28SVAGFVTNSANYY491 pKa = 8.73TPIEE495 pKa = 4.28EE496 pKa = 5.16PYY498 pKa = 11.0LPDD501 pKa = 5.34PNLQPGGGGNPIRR514 pKa = 11.84SSDD517 pKa = 3.54FYY519 pKa = 11.24EE520 pKa = 3.75WNPYY524 pKa = 9.5FDD526 pKa = 5.08EE527 pKa = 6.37LDD529 pKa = 3.38FTTDD533 pKa = 2.31IRR535 pKa = 11.84EE536 pKa = 4.16AFISRR541 pKa = 11.84GMPDD545 pKa = 4.8DD546 pKa = 3.62IGMLIDD552 pKa = 3.56TGRR555 pKa = 11.84NGWGGPDD562 pKa = 3.37RR563 pKa = 11.84PSGVSDD569 pKa = 3.17ATDD572 pKa = 2.85VNTYY576 pKa = 9.92VDD578 pKa = 3.68EE579 pKa = 4.51SRR581 pKa = 11.84VDD583 pKa = 3.59RR584 pKa = 11.84RR585 pKa = 11.84PHH587 pKa = 6.03RR588 pKa = 11.84GGWCNQAGAGIGEE601 pKa = 4.74RR602 pKa = 11.84PTVSPGPGIDD612 pKa = 2.88AYY614 pKa = 11.43VWVKK618 pKa = 10.58PPGEE622 pKa = 4.1SDD624 pKa = 3.68GVSDD628 pKa = 5.66PDD630 pKa = 5.17FEE632 pKa = 6.08PDD634 pKa = 3.95PNDD637 pKa = 3.56PNKK640 pKa = 10.75KK641 pKa = 9.17HH642 pKa = 7.13DD643 pKa = 3.9AMCNPEE649 pKa = 4.19GQSTYY654 pKa = 11.41DD655 pKa = 3.25PDD657 pKa = 4.15YY658 pKa = 8.78PTNALPNAPHH668 pKa = 7.03AGRR671 pKa = 11.84WFPEE675 pKa = 3.73QFAMLVEE682 pKa = 4.11NAYY685 pKa = 9.53PAFF688 pKa = 4.42
MM1 pKa = 7.73RR2 pKa = 11.84FDD4 pKa = 3.34RR5 pKa = 11.84HH6 pKa = 5.79SGRR9 pKa = 11.84HH10 pKa = 4.66RR11 pKa = 11.84LAGASALLAATGLMAGGLIATVATSAHH38 pKa = 6.12AAVACEE44 pKa = 3.86VDD46 pKa = 3.73YY47 pKa = 11.48SANTWTDD54 pKa = 3.35GASSGGFTASVTINNLGDD72 pKa = 3.9PVEE75 pKa = 4.45GWALEE80 pKa = 3.97FEE82 pKa = 4.89LPDD85 pKa = 3.9GQSFTQGWSANWDD98 pKa = 3.44ADD100 pKa = 3.73GTSITAEE107 pKa = 3.71NLAWNGTLGTGASTSVGFNGSWTGSYY133 pKa = 9.77TDD135 pKa = 4.34PSAFTLNGVACTGDD149 pKa = 3.67PGNGGDD155 pKa = 4.17NLPPTVSVTSPSHH168 pKa = 5.01GAEE171 pKa = 3.95YY172 pKa = 10.83DD173 pKa = 3.36EE174 pKa = 5.27GATVPLAATASDD186 pKa = 3.96PDD188 pKa = 3.74GTIDD192 pKa = 3.59RR193 pKa = 11.84VEE195 pKa = 4.05FLVDD199 pKa = 3.31GDD201 pKa = 4.36VVATDD206 pKa = 3.7ASAPYY211 pKa = 9.02EE212 pKa = 4.14ATVSGLDD219 pKa = 3.72TGSHH223 pKa = 4.98TASAVAYY230 pKa = 10.15DD231 pKa = 4.7DD232 pKa = 4.89GDD234 pKa = 4.32PEE236 pKa = 5.27LSATDD241 pKa = 3.56SASFTIGDD249 pKa = 4.6DD250 pKa = 4.1GEE252 pKa = 4.43EE253 pKa = 4.45PPPDD257 pKa = 3.8SDD259 pKa = 4.2NPYY262 pKa = 10.62AGADD266 pKa = 3.57VYY268 pKa = 11.68VNPDD272 pKa = 2.6WAGQVLDD279 pKa = 4.51EE280 pKa = 4.77ASSTGGALGAAMEE293 pKa = 4.49EE294 pKa = 4.69VATYY298 pKa = 7.49PTFVWMDD305 pKa = 4.32RR306 pKa = 11.84IGAITDD312 pKa = 3.33GTGLVGHH319 pKa = 7.63LDD321 pKa = 3.4NAVAQDD327 pKa = 3.71QANGATPTVFQVVIYY342 pKa = 9.25NLPNRR347 pKa = 11.84DD348 pKa = 3.57CAASASNGEE357 pKa = 4.29LLISEE362 pKa = 4.92NGFQRR367 pKa = 11.84YY368 pKa = 8.29RR369 pKa = 11.84EE370 pKa = 4.1EE371 pKa = 4.6YY372 pKa = 8.85IDD374 pKa = 5.06PIAEE378 pKa = 3.75ILSRR382 pKa = 11.84PEE384 pKa = 3.74YY385 pKa = 10.28SDD387 pKa = 4.89LRR389 pKa = 11.84IATVIEE395 pKa = 4.32VDD397 pKa = 3.7SLPNLVTNSDD407 pKa = 4.31DD408 pKa = 4.25PDD410 pKa = 3.86CQEE413 pKa = 3.79AAGDD417 pKa = 3.42GGYY420 pKa = 10.03RR421 pKa = 11.84DD422 pKa = 4.56GIRR425 pKa = 11.84YY426 pKa = 8.89ALNEE430 pKa = 3.89LSAIPNVYY438 pKa = 10.38NYY440 pKa = 10.97LDD442 pKa = 3.68IAHH445 pKa = 6.58SGWLGWDD452 pKa = 3.66SNFGPAVDD460 pKa = 5.74LYY462 pKa = 10.65VQTLTSSDD470 pKa = 3.9FYY472 pKa = 11.69GPAPGFDD479 pKa = 3.28SVAGFVTNSANYY491 pKa = 8.73TPIEE495 pKa = 4.28EE496 pKa = 5.16PYY498 pKa = 11.0LPDD501 pKa = 5.34PNLQPGGGGNPIRR514 pKa = 11.84SSDD517 pKa = 3.54FYY519 pKa = 11.24EE520 pKa = 3.75WNPYY524 pKa = 9.5FDD526 pKa = 5.08EE527 pKa = 6.37LDD529 pKa = 3.38FTTDD533 pKa = 2.31IRR535 pKa = 11.84EE536 pKa = 4.16AFISRR541 pKa = 11.84GMPDD545 pKa = 4.8DD546 pKa = 3.62IGMLIDD552 pKa = 3.56TGRR555 pKa = 11.84NGWGGPDD562 pKa = 3.37RR563 pKa = 11.84PSGVSDD569 pKa = 3.17ATDD572 pKa = 2.85VNTYY576 pKa = 9.92VDD578 pKa = 3.68EE579 pKa = 4.51SRR581 pKa = 11.84VDD583 pKa = 3.59RR584 pKa = 11.84RR585 pKa = 11.84PHH587 pKa = 6.03RR588 pKa = 11.84GGWCNQAGAGIGEE601 pKa = 4.74RR602 pKa = 11.84PTVSPGPGIDD612 pKa = 2.88AYY614 pKa = 11.43VWVKK618 pKa = 10.58PPGEE622 pKa = 4.1SDD624 pKa = 3.68GVSDD628 pKa = 5.66PDD630 pKa = 5.17FEE632 pKa = 6.08PDD634 pKa = 3.95PNDD637 pKa = 3.56PNKK640 pKa = 10.75KK641 pKa = 9.17HH642 pKa = 7.13DD643 pKa = 3.9AMCNPEE649 pKa = 4.19GQSTYY654 pKa = 11.41DD655 pKa = 3.25PDD657 pKa = 4.15YY658 pKa = 8.78PTNALPNAPHH668 pKa = 7.03AGRR671 pKa = 11.84WFPEE675 pKa = 3.73QFAMLVEE682 pKa = 4.11NAYY685 pKa = 9.53PAFF688 pKa = 4.42
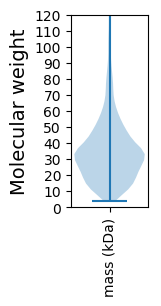
Molecular weight: 72.48 kDa
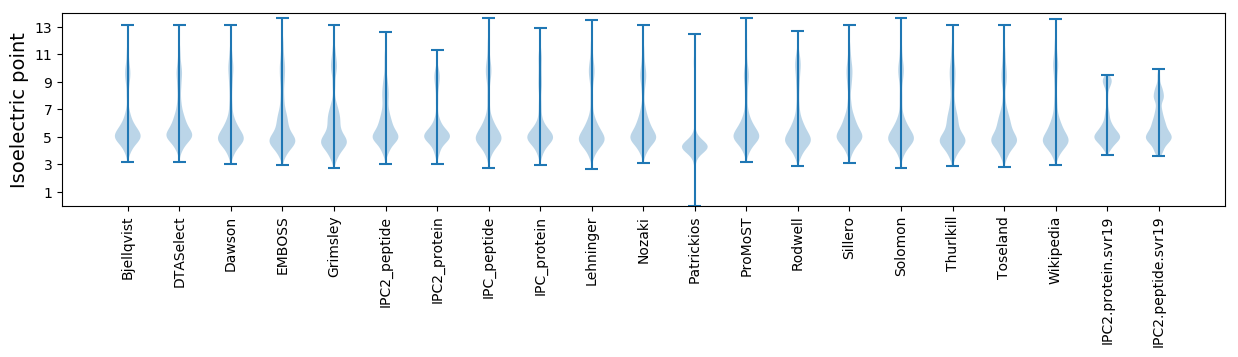
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6L9SDU8|A0A6L9SDU8_9ACTN Peptidoglycan DD-metalloendopeptidase family protein OS=Phytoactinopolyspora halotolerans OX=1981512 GN=G1H10_24395 PE=4 SV=1
MM1 pKa = 7.49ARR3 pKa = 11.84RR4 pKa = 11.84VLGAGWKK11 pKa = 9.42LLARR15 pKa = 11.84RR16 pKa = 11.84VLRR19 pKa = 11.84ARR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84LLAGRR28 pKa = 11.84EE29 pKa = 3.61RR30 pKa = 11.84LLRR33 pKa = 11.84LRR35 pKa = 11.84MRR37 pKa = 11.84KK38 pKa = 6.35TRR40 pKa = 11.84RR41 pKa = 11.84WTVLRR46 pKa = 11.84IPSRR50 pKa = 11.84TVGAGVGVRR59 pKa = 11.84LRR61 pKa = 11.84SGRR64 pKa = 11.84IRR66 pKa = 11.84RR67 pKa = 11.84LFRR70 pKa = 11.84TVGLAGGVWLLRR82 pKa = 11.84RR83 pKa = 11.84LAGG86 pKa = 3.63
MM1 pKa = 7.49ARR3 pKa = 11.84RR4 pKa = 11.84VLGAGWKK11 pKa = 9.42LLARR15 pKa = 11.84RR16 pKa = 11.84VLRR19 pKa = 11.84ARR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84LLAGRR28 pKa = 11.84EE29 pKa = 3.61RR30 pKa = 11.84LLRR33 pKa = 11.84LRR35 pKa = 11.84MRR37 pKa = 11.84KK38 pKa = 6.35TRR40 pKa = 11.84RR41 pKa = 11.84WTVLRR46 pKa = 11.84IPSRR50 pKa = 11.84TVGAGVGVRR59 pKa = 11.84LRR61 pKa = 11.84SGRR64 pKa = 11.84IRR66 pKa = 11.84RR67 pKa = 11.84LFRR70 pKa = 11.84TVGLAGGVWLLRR82 pKa = 11.84RR83 pKa = 11.84LAGG86 pKa = 3.63
Molecular weight: 9.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2132900 |
33 |
3373 |
340.4 |
36.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.701 ± 0.041 | 0.69 ± 0.009 |
6.906 ± 0.028 | 5.986 ± 0.027 |
2.878 ± 0.017 | 9.083 ± 0.028 |
2.29 ± 0.013 | 3.851 ± 0.02 |
1.52 ± 0.018 | 9.799 ± 0.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.927 ± 0.013 | 1.912 ± 0.017 |
5.655 ± 0.02 | 2.807 ± 0.015 |
7.916 ± 0.033 | 5.38 ± 0.018 |
6.004 ± 0.023 | 8.914 ± 0.028 |
1.601 ± 0.016 | 2.178 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
