
Halopenitus malekzadehii
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Haloferacales; Halorubraceae; Halopenitus
Average proteome isoelectric point is 4.9
Get precalculated fractions of proteins
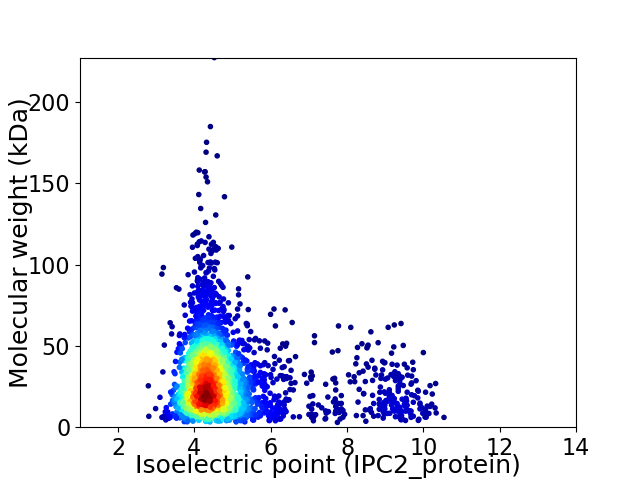
Virtual 2D-PAGE plot for 3119 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
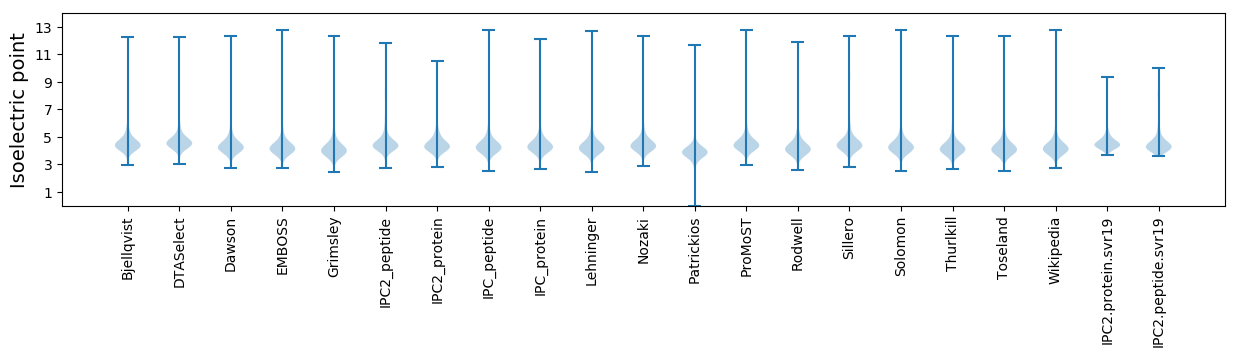
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H6I0M3|A0A1H6I0M3_9EURY Inosose dehydratase OS=Halopenitus malekzadehii OX=1267564 GN=SAMN05192561_101850 PE=4 SV=1
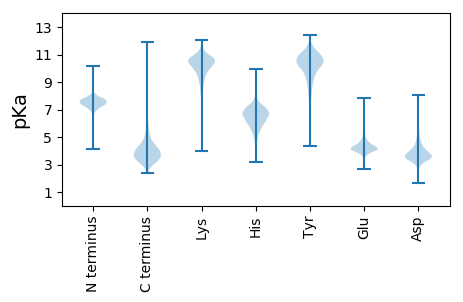
MM1 pKa = 7.44EE2 pKa = 4.86LTWHH6 pKa = 6.23GHH8 pKa = 4.5STWHH12 pKa = 5.64VSVGEE17 pKa = 4.05TDD19 pKa = 4.61LLIDD23 pKa = 4.7PFFDD27 pKa = 3.66NPKK30 pKa = 8.95TDD32 pKa = 3.31VSPEE36 pKa = 4.0EE37 pKa = 4.61LDD39 pKa = 3.43PDD41 pKa = 4.2YY42 pKa = 11.68LLLTHH47 pKa = 6.39GHH49 pKa = 7.12ADD51 pKa = 3.47HH52 pKa = 6.97IGDD55 pKa = 3.38VDD57 pKa = 4.04RR58 pKa = 11.84YY59 pKa = 10.68EE60 pKa = 4.71DD61 pKa = 3.6ATLVATPEE69 pKa = 4.07LVGYY73 pKa = 9.54VQDD76 pKa = 4.05TFGHH80 pKa = 6.67DD81 pKa = 3.44SAVGGMGMNLGGTVEE96 pKa = 4.54CGDD99 pKa = 3.9AFVTMVRR106 pKa = 11.84ADD108 pKa = 3.54HH109 pKa = 6.9SNGIDD114 pKa = 3.12TGYY117 pKa = 8.63GTSAGMPAGFVVSDD131 pKa = 3.9SKK133 pKa = 10.4PTQEE137 pKa = 4.52SDD139 pKa = 3.39AEE141 pKa = 4.34SSTFYY146 pKa = 10.72HH147 pKa = 7.1AGDD150 pKa = 3.45TSLMVEE156 pKa = 4.09MRR158 pKa = 11.84EE159 pKa = 4.17VIGPFLEE166 pKa = 4.65PDD168 pKa = 3.65VAALPAGDD176 pKa = 4.73HH177 pKa = 5.58FTMGPAQAAIAADD190 pKa = 3.54WVGADD195 pKa = 3.4VAIPMHH201 pKa = 6.7YY202 pKa = 10.59DD203 pKa = 3.25SFPPIEE209 pKa = 5.17IDD211 pKa = 2.8TDD213 pKa = 3.77DD214 pKa = 3.75FVRR217 pKa = 11.84EE218 pKa = 4.12VEE220 pKa = 4.28ATAGPSEE227 pKa = 4.09PVVLEE232 pKa = 3.82GDD234 pKa = 3.37EE235 pKa = 4.4SYY237 pKa = 11.15EE238 pKa = 3.83II239 pKa = 4.44
MM1 pKa = 7.44EE2 pKa = 4.86LTWHH6 pKa = 6.23GHH8 pKa = 4.5STWHH12 pKa = 5.64VSVGEE17 pKa = 4.05TDD19 pKa = 4.61LLIDD23 pKa = 4.7PFFDD27 pKa = 3.66NPKK30 pKa = 8.95TDD32 pKa = 3.31VSPEE36 pKa = 4.0EE37 pKa = 4.61LDD39 pKa = 3.43PDD41 pKa = 4.2YY42 pKa = 11.68LLLTHH47 pKa = 6.39GHH49 pKa = 7.12ADD51 pKa = 3.47HH52 pKa = 6.97IGDD55 pKa = 3.38VDD57 pKa = 4.04RR58 pKa = 11.84YY59 pKa = 10.68EE60 pKa = 4.71DD61 pKa = 3.6ATLVATPEE69 pKa = 4.07LVGYY73 pKa = 9.54VQDD76 pKa = 4.05TFGHH80 pKa = 6.67DD81 pKa = 3.44SAVGGMGMNLGGTVEE96 pKa = 4.54CGDD99 pKa = 3.9AFVTMVRR106 pKa = 11.84ADD108 pKa = 3.54HH109 pKa = 6.9SNGIDD114 pKa = 3.12TGYY117 pKa = 8.63GTSAGMPAGFVVSDD131 pKa = 3.9SKK133 pKa = 10.4PTQEE137 pKa = 4.52SDD139 pKa = 3.39AEE141 pKa = 4.34SSTFYY146 pKa = 10.72HH147 pKa = 7.1AGDD150 pKa = 3.45TSLMVEE156 pKa = 4.09MRR158 pKa = 11.84EE159 pKa = 4.17VIGPFLEE166 pKa = 4.65PDD168 pKa = 3.65VAALPAGDD176 pKa = 4.73HH177 pKa = 5.58FTMGPAQAAIAADD190 pKa = 3.54WVGADD195 pKa = 3.4VAIPMHH201 pKa = 6.7YY202 pKa = 10.59DD203 pKa = 3.25SFPPIEE209 pKa = 5.17IDD211 pKa = 2.8TDD213 pKa = 3.77DD214 pKa = 3.75FVRR217 pKa = 11.84EE218 pKa = 4.12VEE220 pKa = 4.28ATAGPSEE227 pKa = 4.09PVVLEE232 pKa = 3.82GDD234 pKa = 3.37EE235 pKa = 4.4SYY237 pKa = 11.15EE238 pKa = 3.83II239 pKa = 4.44
Molecular weight: 25.55 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H6IUY4|A0A1H6IUY4_9EURY Uncharacterized protein OS=Halopenitus malekzadehii OX=1267564 GN=SAMN05192561_103196 PE=4 SV=1
MM1 pKa = 7.03TSVRR5 pKa = 11.84TRR7 pKa = 11.84LALRR11 pKa = 11.84TRR13 pKa = 11.84GNSRR17 pKa = 11.84VTGLKK22 pKa = 10.05RR23 pKa = 11.84IVPAVLSWGIDD34 pKa = 3.06SCLALIVKK42 pKa = 8.25FYY44 pKa = 11.11QRR46 pKa = 11.84VLTGPLFTIIGIEE59 pKa = 3.95RR60 pKa = 11.84LKK62 pKa = 11.21HH63 pKa = 5.57FVAEE67 pKa = 4.68FNQFRR72 pKa = 11.84ISCHH76 pKa = 5.32DD77 pKa = 3.55TVNEE81 pKa = 4.17CVITT85 pKa = 3.9
MM1 pKa = 7.03TSVRR5 pKa = 11.84TRR7 pKa = 11.84LALRR11 pKa = 11.84TRR13 pKa = 11.84GNSRR17 pKa = 11.84VTGLKK22 pKa = 10.05RR23 pKa = 11.84IVPAVLSWGIDD34 pKa = 3.06SCLALIVKK42 pKa = 8.25FYY44 pKa = 11.11QRR46 pKa = 11.84VLTGPLFTIIGIEE59 pKa = 3.95RR60 pKa = 11.84LKK62 pKa = 11.21HH63 pKa = 5.57FVAEE67 pKa = 4.68FNQFRR72 pKa = 11.84ISCHH76 pKa = 5.32DD77 pKa = 3.55TVNEE81 pKa = 4.17CVITT85 pKa = 3.9
Molecular weight: 9.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
899592 |
29 |
2049 |
288.4 |
31.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.071 ± 0.068 | 0.721 ± 0.023 |
8.781 ± 0.062 | 8.18 ± 0.064 |
3.172 ± 0.03 | 8.599 ± 0.041 |
2.051 ± 0.021 | 4.6 ± 0.034 |
1.72 ± 0.028 | 8.588 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.638 ± 0.021 | 2.304 ± 0.029 |
4.766 ± 0.03 | 2.263 ± 0.028 |
6.736 ± 0.046 | 5.485 ± 0.033 |
6.804 ± 0.037 | 8.843 ± 0.049 |
1.084 ± 0.017 | 2.591 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
