
Myroides indicus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Myroides
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
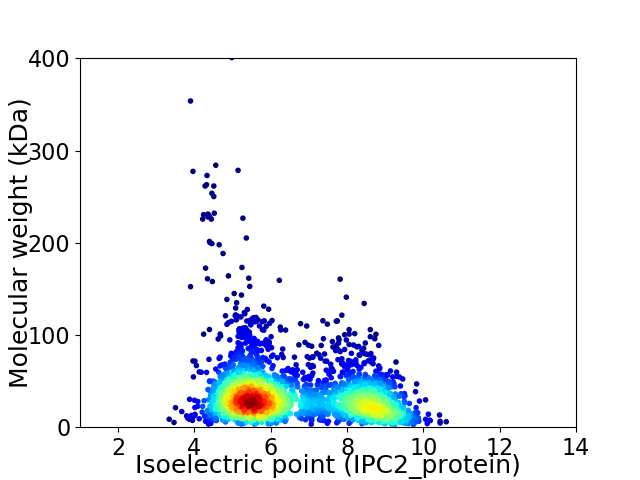
Virtual 2D-PAGE plot for 2689 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R7ETI3|A0A4R7ETI3_9FLAO Cell division protein FtsA OS=Myroides indicus OX=1323422 GN=ftsA PE=3 SV=1
MM1 pKa = 7.9KK2 pKa = 10.18YY3 pKa = 10.7FKK5 pKa = 10.32FLSVFALFGALFLTSCSTDD24 pKa = 3.75DD25 pKa = 5.75KK26 pKa = 10.97DD27 pKa = 4.55TQPTDD32 pKa = 3.29QQIEE36 pKa = 4.37NKK38 pKa = 10.41SFFHH42 pKa = 7.68DD43 pKa = 4.33LFHH46 pKa = 8.06PDD48 pKa = 3.25GLQSNSRR55 pKa = 11.84YY56 pKa = 5.3TTFAAGDD63 pKa = 3.6YY64 pKa = 10.34AGTYY68 pKa = 8.2TGTYY72 pKa = 9.42VINSMLAGSGPGDD85 pKa = 3.6YY86 pKa = 10.97DD87 pKa = 3.73DD88 pKa = 6.35VEE90 pKa = 4.9DD91 pKa = 4.38KK92 pKa = 11.26SITITVNEE100 pKa = 4.19TAQTIDD106 pKa = 4.66LEE108 pKa = 4.23IEE110 pKa = 4.05PFKK113 pKa = 10.85VGSMPANINIEE124 pKa = 4.39MYY126 pKa = 10.3SIPYY130 pKa = 8.81TEE132 pKa = 4.54TGNTVHH138 pKa = 6.71FQGTEE143 pKa = 3.91TNGFSVMGVYY153 pKa = 10.46SDD155 pKa = 3.88IDD157 pKa = 3.5VDD159 pKa = 3.95GTITKK164 pKa = 10.47SGTTYY169 pKa = 10.8TMALNFLANGSWGSMIINADD189 pKa = 3.08VDD191 pKa = 4.26FTGTKK196 pKa = 9.97
MM1 pKa = 7.9KK2 pKa = 10.18YY3 pKa = 10.7FKK5 pKa = 10.32FLSVFALFGALFLTSCSTDD24 pKa = 3.75DD25 pKa = 5.75KK26 pKa = 10.97DD27 pKa = 4.55TQPTDD32 pKa = 3.29QQIEE36 pKa = 4.37NKK38 pKa = 10.41SFFHH42 pKa = 7.68DD43 pKa = 4.33LFHH46 pKa = 8.06PDD48 pKa = 3.25GLQSNSRR55 pKa = 11.84YY56 pKa = 5.3TTFAAGDD63 pKa = 3.6YY64 pKa = 10.34AGTYY68 pKa = 8.2TGTYY72 pKa = 9.42VINSMLAGSGPGDD85 pKa = 3.6YY86 pKa = 10.97DD87 pKa = 3.73DD88 pKa = 6.35VEE90 pKa = 4.9DD91 pKa = 4.38KK92 pKa = 11.26SITITVNEE100 pKa = 4.19TAQTIDD106 pKa = 4.66LEE108 pKa = 4.23IEE110 pKa = 4.05PFKK113 pKa = 10.85VGSMPANINIEE124 pKa = 4.39MYY126 pKa = 10.3SIPYY130 pKa = 8.81TEE132 pKa = 4.54TGNTVHH138 pKa = 6.71FQGTEE143 pKa = 3.91TNGFSVMGVYY153 pKa = 10.46SDD155 pKa = 3.88IDD157 pKa = 3.5VDD159 pKa = 3.95GTITKK164 pKa = 10.47SGTTYY169 pKa = 10.8TMALNFLANGSWGSMIINADD189 pKa = 3.08VDD191 pKa = 4.26FTGTKK196 pKa = 9.97
Molecular weight: 21.43 kDa
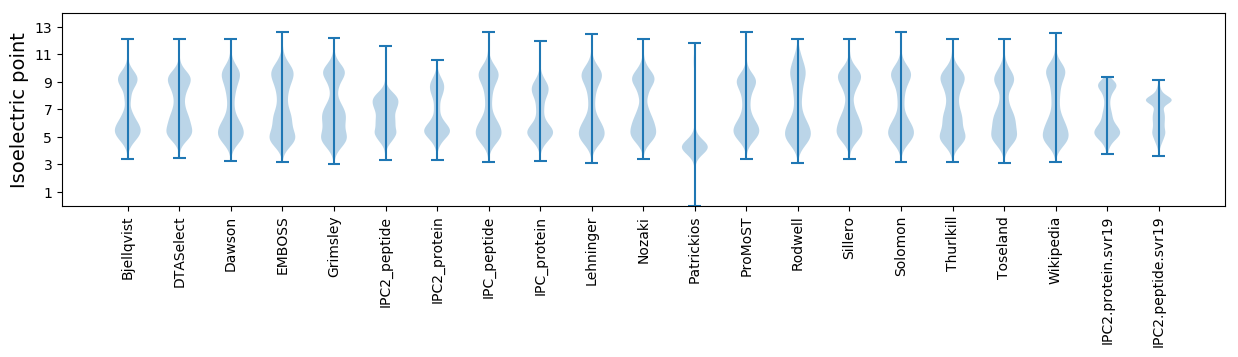
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V3E842|A0A4V3E842_9FLAO RHS repeat-associated protein OS=Myroides indicus OX=1323422 GN=C8P70_11740 PE=4 SV=1
MM1 pKa = 7.36LRR3 pKa = 11.84HH4 pKa = 5.93EE5 pKa = 4.48GRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84TFFHH13 pKa = 6.02NLKK16 pKa = 9.9LASILSTVAGIVNIVGVLSVKK37 pKa = 10.45VLTTNVTGHH46 pKa = 6.39FAFFSEE52 pKa = 4.16EE53 pKa = 4.46LFFKK57 pKa = 10.45DD58 pKa = 3.54YY59 pKa = 11.42SLAGLFLIYY68 pKa = 10.18IFSFLGGAFISSLLMDD84 pKa = 3.99FSKK87 pKa = 10.9RR88 pKa = 11.84IRR90 pKa = 11.84FRR92 pKa = 11.84STYY95 pKa = 8.86ILPVFLEE102 pKa = 3.79ITLFILVMVCSLFYY116 pKa = 10.63TGVDD120 pKa = 3.29QAIASMLLFAMGLQNALVTRR140 pKa = 11.84VSQSVVRR147 pKa = 11.84TTHH150 pKa = 5.1LTGLFTDD157 pKa = 4.66LGIEE161 pKa = 4.21LSFLFFKK168 pKa = 10.2QHH170 pKa = 4.35YY171 pKa = 8.36KK172 pKa = 10.29KK173 pKa = 10.43RR174 pKa = 11.84SLRR177 pKa = 11.84RR178 pKa = 11.84SIGLKK183 pKa = 9.72ISIIGGFFLGCILGGFVYY201 pKa = 10.08QQLEE205 pKa = 4.45IYY207 pKa = 9.77TLFIPVALLLFALWYY222 pKa = 10.16DD223 pKa = 3.55RR224 pKa = 11.84LIFKK228 pKa = 9.92YY229 pKa = 10.69LILRR233 pKa = 11.84RR234 pKa = 11.84KK235 pKa = 10.2LIAQRR240 pKa = 11.84RR241 pKa = 11.84NILKK245 pKa = 10.29LRR247 pKa = 3.89
MM1 pKa = 7.36LRR3 pKa = 11.84HH4 pKa = 5.93EE5 pKa = 4.48GRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84TFFHH13 pKa = 6.02NLKK16 pKa = 9.9LASILSTVAGIVNIVGVLSVKK37 pKa = 10.45VLTTNVTGHH46 pKa = 6.39FAFFSEE52 pKa = 4.16EE53 pKa = 4.46LFFKK57 pKa = 10.45DD58 pKa = 3.54YY59 pKa = 11.42SLAGLFLIYY68 pKa = 10.18IFSFLGGAFISSLLMDD84 pKa = 3.99FSKK87 pKa = 10.9RR88 pKa = 11.84IRR90 pKa = 11.84FRR92 pKa = 11.84STYY95 pKa = 8.86ILPVFLEE102 pKa = 3.79ITLFILVMVCSLFYY116 pKa = 10.63TGVDD120 pKa = 3.29QAIASMLLFAMGLQNALVTRR140 pKa = 11.84VSQSVVRR147 pKa = 11.84TTHH150 pKa = 5.1LTGLFTDD157 pKa = 4.66LGIEE161 pKa = 4.21LSFLFFKK168 pKa = 10.2QHH170 pKa = 4.35YY171 pKa = 8.36KK172 pKa = 10.29KK173 pKa = 10.43RR174 pKa = 11.84SLRR177 pKa = 11.84RR178 pKa = 11.84SIGLKK183 pKa = 9.72ISIIGGFFLGCILGGFVYY201 pKa = 10.08QQLEE205 pKa = 4.45IYY207 pKa = 9.77TLFIPVALLLFALWYY222 pKa = 10.16DD223 pKa = 3.55RR224 pKa = 11.84LIFKK228 pKa = 9.92YY229 pKa = 10.69LILRR233 pKa = 11.84RR234 pKa = 11.84KK235 pKa = 10.2LIAQRR240 pKa = 11.84RR241 pKa = 11.84NILKK245 pKa = 10.29LRR247 pKa = 3.89
Molecular weight: 28.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
898407 |
26 |
3563 |
334.1 |
37.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.078 ± 0.05 | 0.815 ± 0.02 |
5.362 ± 0.035 | 6.873 ± 0.056 |
5.203 ± 0.038 | 6.245 ± 0.047 |
1.712 ± 0.021 | 8.242 ± 0.045 |
7.898 ± 0.057 | 9.236 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.231 ± 0.026 | 6.092 ± 0.044 |
3.248 ± 0.029 | 3.687 ± 0.031 |
3.355 ± 0.033 | 6.413 ± 0.048 |
5.671 ± 0.055 | 6.152 ± 0.04 |
1.043 ± 0.02 | 4.446 ± 0.042 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
