
Polyangium fumosum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Myxococcales; Sorangiineae; Polyangiaceae; Polyangium
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
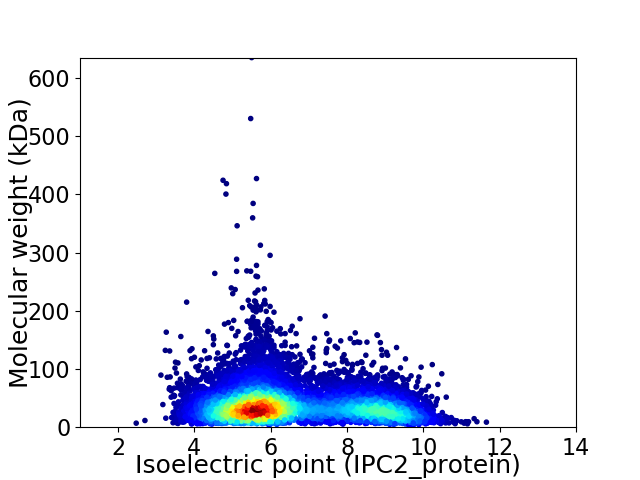
Virtual 2D-PAGE plot for 9827 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
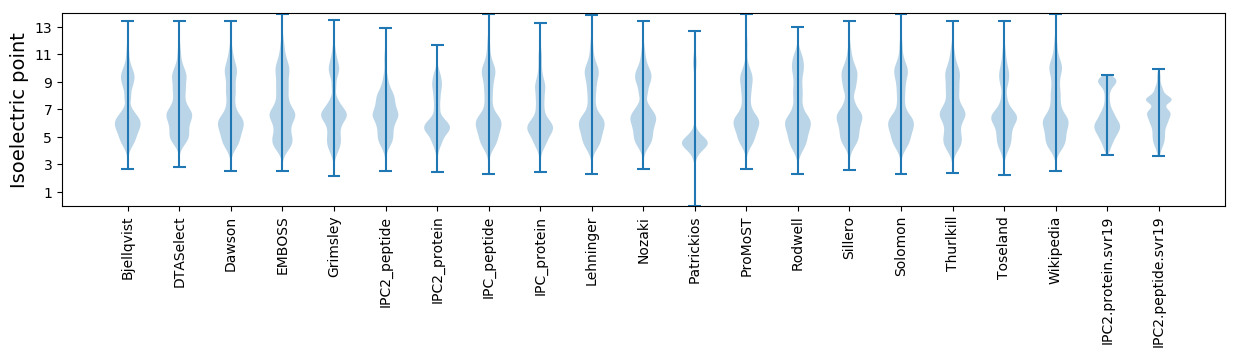
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4U1JGS0|A0A4U1JGS0_9DELT Uncharacterized protein OS=Polyangium fumosum OX=889272 GN=E8A74_06320 PE=4 SV=1
MM1 pKa = 7.27KK2 pKa = 10.35RR3 pKa = 11.84SILVLGLLGTALAACGEE20 pKa = 4.6GAGLPPRR27 pKa = 11.84SNGSSSSTSSSAGPGGAGGQGGEE50 pKa = 4.48GGGGGIGGQGGAGGQGGAGGKK71 pKa = 6.4PTPVCGDD78 pKa = 3.75DD79 pKa = 4.38ALDD82 pKa = 4.19SGEE85 pKa = 4.32EE86 pKa = 4.03CDD88 pKa = 5.78DD89 pKa = 4.09GNTEE93 pKa = 4.6AGDD96 pKa = 4.02GCSPEE101 pKa = 4.26CTVEE105 pKa = 3.94LGEE108 pKa = 5.05IEE110 pKa = 4.98PNDD113 pKa = 4.12TPDD116 pKa = 3.0QASPYY121 pKa = 9.79KK122 pKa = 10.74DD123 pKa = 3.4PFPAKK128 pKa = 9.78IAPEE132 pKa = 4.16GDD134 pKa = 3.0VDD136 pKa = 4.59FVSFTVAATNTSVIARR152 pKa = 11.84ILDD155 pKa = 3.65VGDD158 pKa = 4.12GGCATGVIDD167 pKa = 5.19TILEE171 pKa = 3.83VRR173 pKa = 11.84GADD176 pKa = 3.25GVTVLASDD184 pKa = 4.21DD185 pKa = 3.89DD186 pKa = 4.74AGEE189 pKa = 4.55GACSRR194 pKa = 11.84AVLPSLAPGSYY205 pKa = 9.42FARR208 pKa = 11.84VVASGAAPSPTFLYY222 pKa = 10.3RR223 pKa = 11.84LRR225 pKa = 11.84IDD227 pKa = 3.56QVADD231 pKa = 3.35VCGDD235 pKa = 3.72GQKK238 pKa = 9.04TPAEE242 pKa = 4.28ACDD245 pKa = 5.12DD246 pKa = 4.18GNTDD250 pKa = 4.4AGDD253 pKa = 4.13GCSPTCSIEE262 pKa = 3.9ITEE265 pKa = 4.5TEE267 pKa = 4.42PNDD270 pKa = 3.79TPATANAFVAPWNGVLTPHH289 pKa = 7.09GDD291 pKa = 3.19VDD293 pKa = 4.26VVSVVLSGPASSLTATTTDD312 pKa = 2.96QGTNACAAKK321 pKa = 9.57TLDD324 pKa = 3.92TIVDD328 pKa = 3.67ILAPDD333 pKa = 4.12GTTVLATGDD342 pKa = 4.61DD343 pKa = 3.89IIGNCGSALAQNLTAGTYY361 pKa = 7.69FVRR364 pKa = 11.84MRR366 pKa = 11.84GGSLAGDD373 pKa = 4.06PSPYY377 pKa = 10.11GIQIVIQQ384 pKa = 3.63
MM1 pKa = 7.27KK2 pKa = 10.35RR3 pKa = 11.84SILVLGLLGTALAACGEE20 pKa = 4.6GAGLPPRR27 pKa = 11.84SNGSSSSTSSSAGPGGAGGQGGEE50 pKa = 4.48GGGGGIGGQGGAGGQGGAGGKK71 pKa = 6.4PTPVCGDD78 pKa = 3.75DD79 pKa = 4.38ALDD82 pKa = 4.19SGEE85 pKa = 4.32EE86 pKa = 4.03CDD88 pKa = 5.78DD89 pKa = 4.09GNTEE93 pKa = 4.6AGDD96 pKa = 4.02GCSPEE101 pKa = 4.26CTVEE105 pKa = 3.94LGEE108 pKa = 5.05IEE110 pKa = 4.98PNDD113 pKa = 4.12TPDD116 pKa = 3.0QASPYY121 pKa = 9.79KK122 pKa = 10.74DD123 pKa = 3.4PFPAKK128 pKa = 9.78IAPEE132 pKa = 4.16GDD134 pKa = 3.0VDD136 pKa = 4.59FVSFTVAATNTSVIARR152 pKa = 11.84ILDD155 pKa = 3.65VGDD158 pKa = 4.12GGCATGVIDD167 pKa = 5.19TILEE171 pKa = 3.83VRR173 pKa = 11.84GADD176 pKa = 3.25GVTVLASDD184 pKa = 4.21DD185 pKa = 3.89DD186 pKa = 4.74AGEE189 pKa = 4.55GACSRR194 pKa = 11.84AVLPSLAPGSYY205 pKa = 9.42FARR208 pKa = 11.84VVASGAAPSPTFLYY222 pKa = 10.3RR223 pKa = 11.84LRR225 pKa = 11.84IDD227 pKa = 3.56QVADD231 pKa = 3.35VCGDD235 pKa = 3.72GQKK238 pKa = 9.04TPAEE242 pKa = 4.28ACDD245 pKa = 5.12DD246 pKa = 4.18GNTDD250 pKa = 4.4AGDD253 pKa = 4.13GCSPTCSIEE262 pKa = 3.9ITEE265 pKa = 4.5TEE267 pKa = 4.42PNDD270 pKa = 3.79TPATANAFVAPWNGVLTPHH289 pKa = 7.09GDD291 pKa = 3.19VDD293 pKa = 4.26VVSVVLSGPASSLTATTTDD312 pKa = 2.96QGTNACAAKK321 pKa = 9.57TLDD324 pKa = 3.92TIVDD328 pKa = 3.67ILAPDD333 pKa = 4.12GTTVLATGDD342 pKa = 4.61DD343 pKa = 3.89IIGNCGSALAQNLTAGTYY361 pKa = 7.69FVRR364 pKa = 11.84MRR366 pKa = 11.84GGSLAGDD373 pKa = 4.06PSPYY377 pKa = 10.11GIQIVIQQ384 pKa = 3.63
Molecular weight: 37.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4U1JJF1|A0A4U1JJF1_9DELT Uncharacterized protein OS=Polyangium fumosum OX=889272 GN=E8A74_03665 PE=4 SV=1
MM1 pKa = 7.5KK2 pKa = 9.41MWTWRR7 pKa = 11.84PRR9 pKa = 11.84GPGRR13 pKa = 11.84RR14 pKa = 11.84PRR16 pKa = 11.84GPGRR20 pKa = 11.84RR21 pKa = 11.84PRR23 pKa = 11.84GPGRR27 pKa = 11.84RR28 pKa = 11.84PRR30 pKa = 11.84GPGRR34 pKa = 11.84RR35 pKa = 11.84PRR37 pKa = 11.84GWRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84PRR44 pKa = 11.84GPGRR48 pKa = 11.84RR49 pKa = 11.84PRR51 pKa = 11.84GWRR54 pKa = 11.84GRR56 pKa = 11.84PRR58 pKa = 11.84GPGRR62 pKa = 11.84RR63 pKa = 11.84PRR65 pKa = 11.84GWRR68 pKa = 11.84GRR70 pKa = 11.84PRR72 pKa = 11.84GSGKK76 pKa = 10.44ASTRR80 pKa = 11.84LEE82 pKa = 3.97RR83 pKa = 11.84ASARR87 pKa = 11.84SGKK90 pKa = 10.48ASTRR94 pKa = 11.84FGKK97 pKa = 10.61ASTRR101 pKa = 11.84FGKK104 pKa = 10.56ASTRR108 pKa = 11.84FEE110 pKa = 3.98RR111 pKa = 11.84ASTRR115 pKa = 11.84FGKK118 pKa = 10.66APTRR122 pKa = 11.84SGKK125 pKa = 10.61ASTRR129 pKa = 3.5
MM1 pKa = 7.5KK2 pKa = 9.41MWTWRR7 pKa = 11.84PRR9 pKa = 11.84GPGRR13 pKa = 11.84RR14 pKa = 11.84PRR16 pKa = 11.84GPGRR20 pKa = 11.84RR21 pKa = 11.84PRR23 pKa = 11.84GPGRR27 pKa = 11.84RR28 pKa = 11.84PRR30 pKa = 11.84GPGRR34 pKa = 11.84RR35 pKa = 11.84PRR37 pKa = 11.84GWRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84PRR44 pKa = 11.84GPGRR48 pKa = 11.84RR49 pKa = 11.84PRR51 pKa = 11.84GWRR54 pKa = 11.84GRR56 pKa = 11.84PRR58 pKa = 11.84GPGRR62 pKa = 11.84RR63 pKa = 11.84PRR65 pKa = 11.84GWRR68 pKa = 11.84GRR70 pKa = 11.84PRR72 pKa = 11.84GSGKK76 pKa = 10.44ASTRR80 pKa = 11.84LEE82 pKa = 3.97RR83 pKa = 11.84ASARR87 pKa = 11.84SGKK90 pKa = 10.48ASTRR94 pKa = 11.84FGKK97 pKa = 10.61ASTRR101 pKa = 11.84FGKK104 pKa = 10.56ASTRR108 pKa = 11.84FEE110 pKa = 3.98RR111 pKa = 11.84ASTRR115 pKa = 11.84FGKK118 pKa = 10.66APTRR122 pKa = 11.84SGKK125 pKa = 10.61ASTRR129 pKa = 3.5
Molecular weight: 14.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3775618 |
28 |
5848 |
384.2 |
41.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.897 ± 0.037 | 1.261 ± 0.019 |
5.68 ± 0.019 | 6.366 ± 0.027 |
3.302 ± 0.013 | 9.005 ± 0.032 |
2.049 ± 0.01 | 3.72 ± 0.015 |
3.145 ± 0.022 | 9.951 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.845 ± 0.01 | 2.052 ± 0.016 |
6.398 ± 0.031 | 2.617 ± 0.012 |
7.941 ± 0.035 | 5.44 ± 0.015 |
5.263 ± 0.02 | 7.811 ± 0.02 |
1.255 ± 0.009 | 1.999 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
