
Desulfomicrobium baculatum (strain DSM 4028 / VKM B-1378 / X) (Desulfovibrio baculatus)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfovibrionales; Desulfomicrobiaceae; Desulfomicrobium; Desulfomicrobium baculatum
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
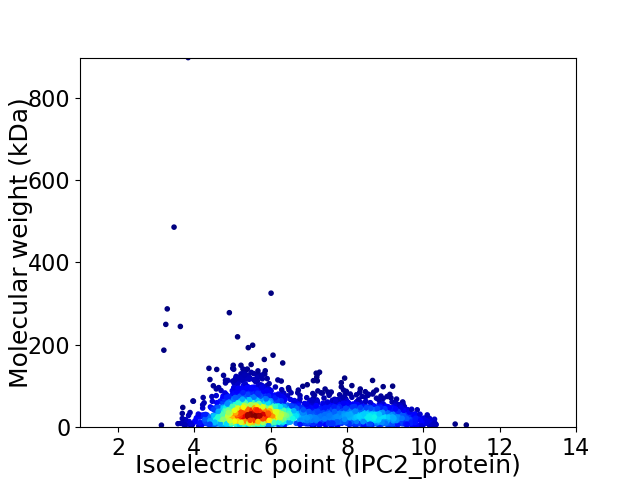
Virtual 2D-PAGE plot for 3421 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C7LVB6|C7LVB6_DESBD Uncharacterized protein OS=Desulfomicrobium baculatum (strain DSM 4028 / VKM B-1378 / X) OX=525897 GN=Dbac_0331 PE=4 SV=1
MM1 pKa = 7.72KK2 pKa = 10.55RR3 pKa = 11.84FTLVALVAALLFSMVTSASATEE25 pKa = 4.01LKK27 pKa = 10.8VRR29 pKa = 11.84GNFDD33 pKa = 3.51VYY35 pKa = 11.44GMWSSNLNDD44 pKa = 4.22HH45 pKa = 7.31DD46 pKa = 5.53SDD48 pKa = 4.33VADD51 pKa = 3.87GDD53 pKa = 3.91NYY55 pKa = 8.89TTVQRR60 pKa = 11.84MRR62 pKa = 11.84TYY64 pKa = 10.51FDD66 pKa = 3.36YY67 pKa = 11.23VANEE71 pKa = 3.76NLKK74 pKa = 10.69AVLGFEE80 pKa = 4.85FDD82 pKa = 4.17NVWGAGEE89 pKa = 3.95EE90 pKa = 4.51ANWGTDD96 pKa = 2.95GKK98 pKa = 9.41EE99 pKa = 3.98TLEE102 pKa = 4.21VKK104 pKa = 10.08HH105 pKa = 6.72AYY107 pKa = 10.52LDD109 pKa = 4.49FIFPDD114 pKa = 3.66TQIEE118 pKa = 4.4VQAGLQYY125 pKa = 10.6IALPSIFGNPVFDD138 pKa = 5.84DD139 pKa = 3.88DD140 pKa = 4.2AAAITVAAPINDD152 pKa = 3.61MFGVTVGYY160 pKa = 8.61TRR162 pKa = 11.84GTDD165 pKa = 3.4MSNSFSDD172 pKa = 3.66EE173 pKa = 4.08LLVSEE178 pKa = 6.2GYY180 pKa = 10.79DD181 pKa = 3.61KK182 pKa = 11.61DD183 pKa = 4.08DD184 pKa = 3.42VDD186 pKa = 3.96MAFIATPVTLDD197 pKa = 3.56GVSVTPYY204 pKa = 10.25FGYY207 pKa = 11.0AWLGQNSTKK216 pKa = 10.8SFDD219 pKa = 4.1ANDD222 pKa = 4.25DD223 pKa = 3.61STVWVVGANAALTMFDD239 pKa = 4.55PLTFAADD246 pKa = 4.66LIYY249 pKa = 11.24GEE251 pKa = 4.97GDD253 pKa = 2.87SDD255 pKa = 4.34AYY257 pKa = 8.46EE258 pKa = 4.2TKK260 pKa = 10.24GWYY263 pKa = 9.24AALAASYY270 pKa = 10.97KK271 pKa = 9.73MDD273 pKa = 3.85MLTATLFTTYY283 pKa = 9.87ATGYY287 pKa = 10.98DD288 pKa = 4.05EE289 pKa = 6.3DD290 pKa = 4.23TDD292 pKa = 4.46EE293 pKa = 5.6DD294 pKa = 4.38DD295 pKa = 5.31KK296 pKa = 11.91LPTLAEE302 pKa = 4.33GWGLTPYY309 pKa = 10.65VGGDD313 pKa = 3.08RR314 pKa = 11.84AFIAANDD321 pKa = 3.97SFGTDD326 pKa = 3.02ALGVGNDD333 pKa = 3.92GTGLWVLGLVLDD345 pKa = 5.88DD346 pKa = 4.27ISFVDD351 pKa = 3.78KK352 pKa = 10.92LSHH355 pKa = 6.52KK356 pKa = 10.58LVIAYY361 pKa = 8.82AQGTSDD367 pKa = 4.15EE368 pKa = 4.91DD369 pKa = 3.38ATKK372 pKa = 10.5PSGAAVKK379 pKa = 8.83FTEE382 pKa = 5.49EE383 pKa = 3.96DD384 pKa = 3.9DD385 pKa = 4.86LWEE388 pKa = 4.01LWLVNKK394 pKa = 9.99YY395 pKa = 9.63MIYY398 pKa = 10.86EE399 pKa = 3.87NLAAINEE406 pKa = 4.24LGYY409 pKa = 10.15FQASSEE415 pKa = 4.4VYY417 pKa = 10.46EE418 pKa = 4.91DD419 pKa = 3.49ATGDD423 pKa = 4.26DD424 pKa = 4.29LDD426 pKa = 3.73SSYY429 pKa = 11.03FATVGFQYY437 pKa = 11.1KK438 pKa = 9.31FF439 pKa = 3.23
MM1 pKa = 7.72KK2 pKa = 10.55RR3 pKa = 11.84FTLVALVAALLFSMVTSASATEE25 pKa = 4.01LKK27 pKa = 10.8VRR29 pKa = 11.84GNFDD33 pKa = 3.51VYY35 pKa = 11.44GMWSSNLNDD44 pKa = 4.22HH45 pKa = 7.31DD46 pKa = 5.53SDD48 pKa = 4.33VADD51 pKa = 3.87GDD53 pKa = 3.91NYY55 pKa = 8.89TTVQRR60 pKa = 11.84MRR62 pKa = 11.84TYY64 pKa = 10.51FDD66 pKa = 3.36YY67 pKa = 11.23VANEE71 pKa = 3.76NLKK74 pKa = 10.69AVLGFEE80 pKa = 4.85FDD82 pKa = 4.17NVWGAGEE89 pKa = 3.95EE90 pKa = 4.51ANWGTDD96 pKa = 2.95GKK98 pKa = 9.41EE99 pKa = 3.98TLEE102 pKa = 4.21VKK104 pKa = 10.08HH105 pKa = 6.72AYY107 pKa = 10.52LDD109 pKa = 4.49FIFPDD114 pKa = 3.66TQIEE118 pKa = 4.4VQAGLQYY125 pKa = 10.6IALPSIFGNPVFDD138 pKa = 5.84DD139 pKa = 3.88DD140 pKa = 4.2AAAITVAAPINDD152 pKa = 3.61MFGVTVGYY160 pKa = 8.61TRR162 pKa = 11.84GTDD165 pKa = 3.4MSNSFSDD172 pKa = 3.66EE173 pKa = 4.08LLVSEE178 pKa = 6.2GYY180 pKa = 10.79DD181 pKa = 3.61KK182 pKa = 11.61DD183 pKa = 4.08DD184 pKa = 3.42VDD186 pKa = 3.96MAFIATPVTLDD197 pKa = 3.56GVSVTPYY204 pKa = 10.25FGYY207 pKa = 11.0AWLGQNSTKK216 pKa = 10.8SFDD219 pKa = 4.1ANDD222 pKa = 4.25DD223 pKa = 3.61STVWVVGANAALTMFDD239 pKa = 4.55PLTFAADD246 pKa = 4.66LIYY249 pKa = 11.24GEE251 pKa = 4.97GDD253 pKa = 2.87SDD255 pKa = 4.34AYY257 pKa = 8.46EE258 pKa = 4.2TKK260 pKa = 10.24GWYY263 pKa = 9.24AALAASYY270 pKa = 10.97KK271 pKa = 9.73MDD273 pKa = 3.85MLTATLFTTYY283 pKa = 9.87ATGYY287 pKa = 10.98DD288 pKa = 4.05EE289 pKa = 6.3DD290 pKa = 4.23TDD292 pKa = 4.46EE293 pKa = 5.6DD294 pKa = 4.38DD295 pKa = 5.31KK296 pKa = 11.91LPTLAEE302 pKa = 4.33GWGLTPYY309 pKa = 10.65VGGDD313 pKa = 3.08RR314 pKa = 11.84AFIAANDD321 pKa = 3.97SFGTDD326 pKa = 3.02ALGVGNDD333 pKa = 3.92GTGLWVLGLVLDD345 pKa = 5.88DD346 pKa = 4.27ISFVDD351 pKa = 3.78KK352 pKa = 10.92LSHH355 pKa = 6.52KK356 pKa = 10.58LVIAYY361 pKa = 8.82AQGTSDD367 pKa = 4.15EE368 pKa = 4.91DD369 pKa = 3.38ATKK372 pKa = 10.5PSGAAVKK379 pKa = 8.83FTEE382 pKa = 5.49EE383 pKa = 3.96DD384 pKa = 3.9DD385 pKa = 4.86LWEE388 pKa = 4.01LWLVNKK394 pKa = 9.99YY395 pKa = 9.63MIYY398 pKa = 10.86EE399 pKa = 3.87NLAAINEE406 pKa = 4.24LGYY409 pKa = 10.15FQASSEE415 pKa = 4.4VYY417 pKa = 10.46EE418 pKa = 4.91DD419 pKa = 3.49ATGDD423 pKa = 4.26DD424 pKa = 4.29LDD426 pKa = 3.73SSYY429 pKa = 11.03FATVGFQYY437 pKa = 11.1KK438 pKa = 9.31FF439 pKa = 3.23
Molecular weight: 47.99 kDa
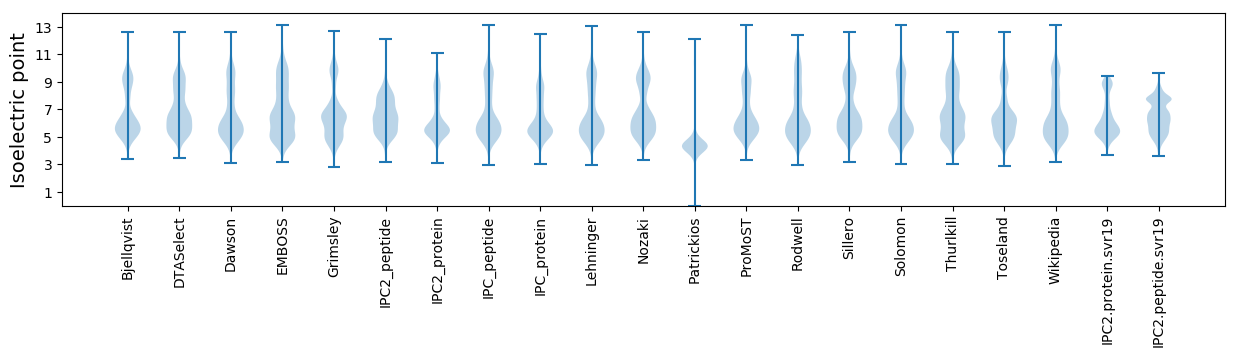
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C7LXK2|C7LXK2_DESBD Uncharacterized protein OS=Desulfomicrobium baculatum (strain DSM 4028 / VKM B-1378 / X) OX=525897 GN=Dbac_3163 PE=4 SV=1
MM1 pKa = 7.74SKK3 pKa = 8.94RR4 pKa = 11.84TYY6 pKa = 10.32QPSTTKK12 pKa = 10.4RR13 pKa = 11.84KK14 pKa = 9.13RR15 pKa = 11.84SHH17 pKa = 6.09GFLVRR22 pKa = 11.84SRR24 pKa = 11.84TKK26 pKa = 10.46NGQAVLRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.65GRR40 pKa = 11.84KK41 pKa = 8.66RR42 pKa = 11.84LAVV45 pKa = 3.41
MM1 pKa = 7.74SKK3 pKa = 8.94RR4 pKa = 11.84TYY6 pKa = 10.32QPSTTKK12 pKa = 10.4RR13 pKa = 11.84KK14 pKa = 9.13RR15 pKa = 11.84SHH17 pKa = 6.09GFLVRR22 pKa = 11.84SRR24 pKa = 11.84TKK26 pKa = 10.46NGQAVLRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.65GRR40 pKa = 11.84KK41 pKa = 8.66RR42 pKa = 11.84LAVV45 pKa = 3.41
Molecular weight: 5.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1171290 |
30 |
8980 |
342.4 |
37.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.738 ± 0.051 | 1.322 ± 0.023 |
5.524 ± 0.063 | 6.277 ± 0.042 |
4.126 ± 0.034 | 8.106 ± 0.079 |
2.181 ± 0.021 | 5.548 ± 0.039 |
4.259 ± 0.044 | 10.924 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.764 ± 0.025 | 3.024 ± 0.033 |
4.632 ± 0.041 | 3.416 ± 0.023 |
6.237 ± 0.053 | 5.856 ± 0.05 |
5.106 ± 0.046 | 7.258 ± 0.041 |
1.196 ± 0.018 | 2.505 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
