
Sunn-hemp mosaic virus (SHMV) (TMV strain cowpea)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Virgaviridae; Tobamovirus
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
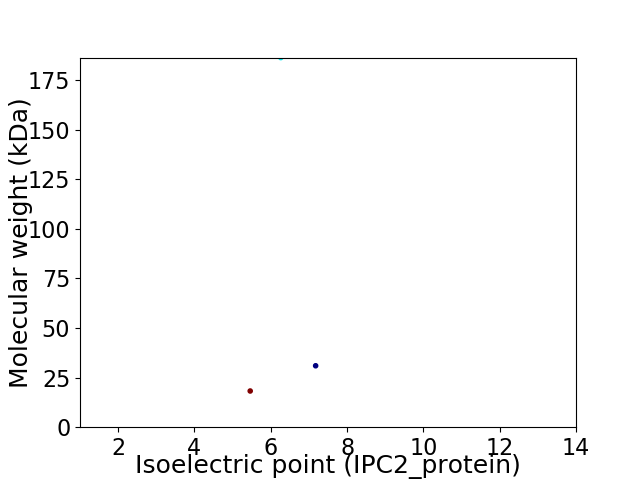
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P03585|MVP_SHMV Movement protein OS=Sunn-hemp mosaic virus OX=12240 GN=MP PE=3 SV=1
MM1 pKa = 7.8AYY3 pKa = 10.35SIPTPSQLVYY13 pKa = 9.43FTEE16 pKa = 4.52NYY18 pKa = 10.25ADD20 pKa = 4.24YY21 pKa = 10.83IPFVNRR27 pKa = 11.84LINARR32 pKa = 11.84SNSFQTQSGRR42 pKa = 11.84DD43 pKa = 3.71EE44 pKa = 4.06LRR46 pKa = 11.84EE47 pKa = 3.87ILIKK51 pKa = 10.83SQVSVVSPISRR62 pKa = 11.84FPAEE66 pKa = 3.76PAYY69 pKa = 10.4YY70 pKa = 9.59IYY72 pKa = 10.95LRR74 pKa = 11.84DD75 pKa = 3.67PSISTVYY82 pKa = 9.51TALLQSTDD90 pKa = 2.58TRR92 pKa = 11.84NRR94 pKa = 11.84VIEE97 pKa = 4.2VEE99 pKa = 3.75NSTNVTTAEE108 pKa = 3.73QLNAVRR114 pKa = 11.84RR115 pKa = 11.84TDD117 pKa = 3.53DD118 pKa = 3.29ASTAIHH124 pKa = 6.63NNLEE128 pKa = 4.1QLLSLLTNGTGVFNRR143 pKa = 11.84TSFEE147 pKa = 4.24SASGLTWLVTTTPRR161 pKa = 11.84TAA163 pKa = 3.4
MM1 pKa = 7.8AYY3 pKa = 10.35SIPTPSQLVYY13 pKa = 9.43FTEE16 pKa = 4.52NYY18 pKa = 10.25ADD20 pKa = 4.24YY21 pKa = 10.83IPFVNRR27 pKa = 11.84LINARR32 pKa = 11.84SNSFQTQSGRR42 pKa = 11.84DD43 pKa = 3.71EE44 pKa = 4.06LRR46 pKa = 11.84EE47 pKa = 3.87ILIKK51 pKa = 10.83SQVSVVSPISRR62 pKa = 11.84FPAEE66 pKa = 3.76PAYY69 pKa = 10.4YY70 pKa = 9.59IYY72 pKa = 10.95LRR74 pKa = 11.84DD75 pKa = 3.67PSISTVYY82 pKa = 9.51TALLQSTDD90 pKa = 2.58TRR92 pKa = 11.84NRR94 pKa = 11.84VIEE97 pKa = 4.2VEE99 pKa = 3.75NSTNVTTAEE108 pKa = 3.73QLNAVRR114 pKa = 11.84RR115 pKa = 11.84TDD117 pKa = 3.53DD118 pKa = 3.29ASTAIHH124 pKa = 6.63NNLEE128 pKa = 4.1QLLSLLTNGTGVFNRR143 pKa = 11.84TSFEE147 pKa = 4.24SASGLTWLVTTTPRR161 pKa = 11.84TAA163 pKa = 3.4
Molecular weight: 18.25 kDa
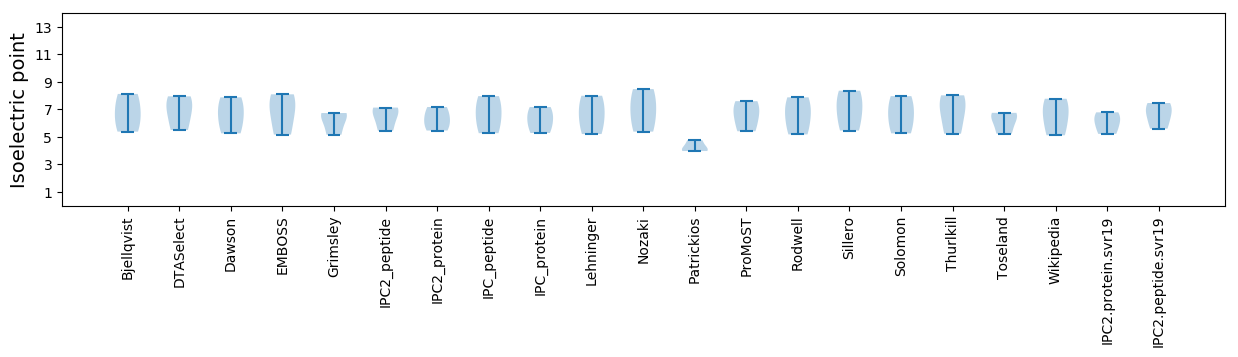
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P89202|RDRP_SHMV Replicase large subunit OS=Sunn-hemp mosaic virus OX=12240 PE=2 SV=2
MM1 pKa = 7.69SEE3 pKa = 3.81VSKK6 pKa = 10.83ISTLLAPEE14 pKa = 4.51KK15 pKa = 10.13FVKK18 pKa = 10.67LSVSDD23 pKa = 3.52KK24 pKa = 10.58FKK26 pKa = 10.06WKK28 pKa = 10.42APSRR32 pKa = 11.84VCSIVQSDD40 pKa = 4.8TISMTANGRR49 pKa = 11.84SLFTFDD55 pKa = 4.13VLKK58 pKa = 10.92DD59 pKa = 3.75VLKK62 pKa = 10.65HH63 pKa = 5.86AEE65 pKa = 4.06EE66 pKa = 4.14YY67 pKa = 10.43TYY69 pKa = 11.52VDD71 pKa = 2.99VLGVVLSGQWLLPKK85 pKa = 8.94GTPGSAEE92 pKa = 4.0IILLDD97 pKa = 3.96SRR99 pKa = 11.84LKK101 pKa = 11.04GKK103 pKa = 10.68ASVLAVFNCRR113 pKa = 11.84AATQEE118 pKa = 4.02FQFLISPGYY127 pKa = 10.79SLTCADD133 pKa = 3.92ALKK136 pKa = 10.68KK137 pKa = 10.39PFEE140 pKa = 4.26ISCNVIDD147 pKa = 5.87LPVKK151 pKa = 10.82DD152 pKa = 4.38GFTPLSVEE160 pKa = 4.13IACLVQFSNCVITRR174 pKa = 11.84SLTMKK179 pKa = 10.46LKK181 pKa = 10.33EE182 pKa = 4.18NPATRR187 pKa = 11.84TFSAEE192 pKa = 4.09EE193 pKa = 3.79VDD195 pKa = 4.06EE196 pKa = 4.75LLGSMTTLRR205 pKa = 11.84SIEE208 pKa = 3.91GLRR211 pKa = 11.84KK212 pKa = 9.72KK213 pKa = 10.57KK214 pKa = 10.34EE215 pKa = 4.06PNDD218 pKa = 3.42VVQGHH223 pKa = 7.03LSAEE227 pKa = 4.15YY228 pKa = 10.13DD229 pKa = 3.45VKK231 pKa = 11.04RR232 pKa = 11.84SVKK235 pKa = 8.92RR236 pKa = 11.84TKK238 pKa = 10.71SEE240 pKa = 3.67NTPGKK245 pKa = 10.15RR246 pKa = 11.84RR247 pKa = 11.84VNVDD251 pKa = 3.42SVSLGLGKK259 pKa = 10.6GKK261 pKa = 10.35SVSAKK266 pKa = 10.73NEE268 pKa = 3.86DD269 pKa = 3.94TEE271 pKa = 5.33SVFDD275 pKa = 5.93DD276 pKa = 5.52GILDD280 pKa = 3.69SDD282 pKa = 4.05SS283 pKa = 3.32
MM1 pKa = 7.69SEE3 pKa = 3.81VSKK6 pKa = 10.83ISTLLAPEE14 pKa = 4.51KK15 pKa = 10.13FVKK18 pKa = 10.67LSVSDD23 pKa = 3.52KK24 pKa = 10.58FKK26 pKa = 10.06WKK28 pKa = 10.42APSRR32 pKa = 11.84VCSIVQSDD40 pKa = 4.8TISMTANGRR49 pKa = 11.84SLFTFDD55 pKa = 4.13VLKK58 pKa = 10.92DD59 pKa = 3.75VLKK62 pKa = 10.65HH63 pKa = 5.86AEE65 pKa = 4.06EE66 pKa = 4.14YY67 pKa = 10.43TYY69 pKa = 11.52VDD71 pKa = 2.99VLGVVLSGQWLLPKK85 pKa = 8.94GTPGSAEE92 pKa = 4.0IILLDD97 pKa = 3.96SRR99 pKa = 11.84LKK101 pKa = 11.04GKK103 pKa = 10.68ASVLAVFNCRR113 pKa = 11.84AATQEE118 pKa = 4.02FQFLISPGYY127 pKa = 10.79SLTCADD133 pKa = 3.92ALKK136 pKa = 10.68KK137 pKa = 10.39PFEE140 pKa = 4.26ISCNVIDD147 pKa = 5.87LPVKK151 pKa = 10.82DD152 pKa = 4.38GFTPLSVEE160 pKa = 4.13IACLVQFSNCVITRR174 pKa = 11.84SLTMKK179 pKa = 10.46LKK181 pKa = 10.33EE182 pKa = 4.18NPATRR187 pKa = 11.84TFSAEE192 pKa = 4.09EE193 pKa = 3.79VDD195 pKa = 4.06EE196 pKa = 4.75LLGSMTTLRR205 pKa = 11.84SIEE208 pKa = 3.91GLRR211 pKa = 11.84KK212 pKa = 9.72KK213 pKa = 10.57KK214 pKa = 10.34EE215 pKa = 4.06PNDD218 pKa = 3.42VVQGHH223 pKa = 7.03LSAEE227 pKa = 4.15YY228 pKa = 10.13DD229 pKa = 3.45VKK231 pKa = 11.04RR232 pKa = 11.84SVKK235 pKa = 8.92RR236 pKa = 11.84TKK238 pKa = 10.71SEE240 pKa = 3.67NTPGKK245 pKa = 10.15RR246 pKa = 11.84RR247 pKa = 11.84VNVDD251 pKa = 3.42SVSLGLGKK259 pKa = 10.6GKK261 pKa = 10.35SVSAKK266 pKa = 10.73NEE268 pKa = 3.86DD269 pKa = 3.94TEE271 pKa = 5.33SVFDD275 pKa = 5.93DD276 pKa = 5.52GILDD280 pKa = 3.69SDD282 pKa = 4.05SS283 pKa = 3.32
Molecular weight: 30.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2075 |
163 |
1629 |
691.7 |
78.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.639 ± 0.434 | 2.072 ± 0.511 |
6.506 ± 0.732 | 5.687 ± 0.192 |
5.108 ± 0.552 | 4.771 ± 0.493 |
1.928 ± 0.622 | 5.494 ± 0.336 |
6.458 ± 1.243 | 9.783 ± 0.217 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.88 ± 0.413 | 4.819 ± 0.568 |
4.096 ± 0.177 | 3.277 ± 0.313 |
5.446 ± 0.425 | 7.566 ± 1.892 |
6.12 ± 1.581 | 8.193 ± 0.457 |
1.06 ± 0.195 | 4.0 ± 0.715 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
