
Snakehead rhabdovirus (SHRV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Gammarhabdovirinae; Novirhabdovirus; Snakehead novirhabdovirus
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
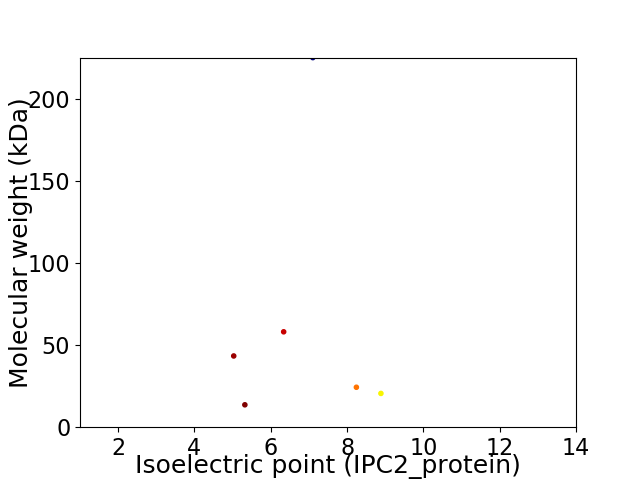
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
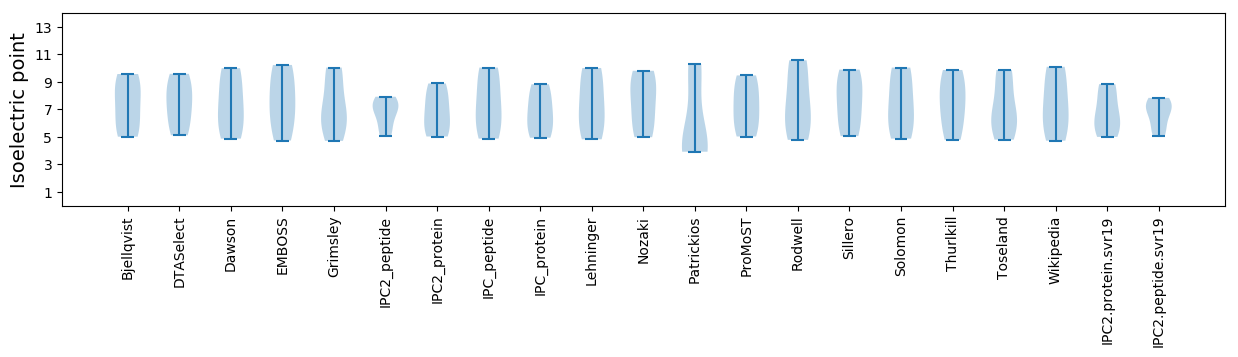
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>sp|Q9QJT9|NCAP_SHRV Nucleoprotein OS=Snakehead rhabdovirus OX=103603 GN=N PE=3 SV=1
MM1 pKa = 7.77AFQKK5 pKa = 10.68EE6 pKa = 4.4FFGLRR11 pKa = 11.84DD12 pKa = 3.59VKK14 pKa = 11.12VDD16 pKa = 3.78LSAGEE21 pKa = 4.52GLDD24 pKa = 3.67FDD26 pKa = 4.51PSEE29 pKa = 4.52VEE31 pKa = 3.73LTVYY35 pKa = 9.26RR36 pKa = 11.84TGADD40 pKa = 3.17TDD42 pKa = 3.77GNTIIKK48 pKa = 10.2ALAAVGGPKK57 pKa = 9.72TNEE60 pKa = 3.79ALSVLLAFVTLGTDD74 pKa = 2.85QDD76 pKa = 3.88EE77 pKa = 4.66YY78 pKa = 9.81EE79 pKa = 4.19TRR81 pKa = 11.84IKK83 pKa = 10.12ILKK86 pKa = 9.84EE87 pKa = 3.14IGFSVKK93 pKa = 9.34EE94 pKa = 3.77VPMAKK99 pKa = 9.87DD100 pKa = 3.06ASSGIEE106 pKa = 3.71APLEE110 pKa = 3.88NVAALVKK117 pKa = 10.53PEE119 pKa = 3.92TVYY122 pKa = 10.85QVIRR126 pKa = 11.84GVLYY130 pKa = 9.55TCALFVKK137 pKa = 10.28YY138 pKa = 10.15NVEE141 pKa = 4.1KK142 pKa = 8.09MQKK145 pKa = 9.45YY146 pKa = 9.75IKK148 pKa = 10.5NKK150 pKa = 10.08LPALATSYY158 pKa = 10.53GVPEE162 pKa = 4.58LEE164 pKa = 4.23EE165 pKa = 4.53FPTEE169 pKa = 3.89SSALKK174 pKa = 10.48KK175 pKa = 10.17LASCIRR181 pKa = 11.84PGQRR185 pKa = 11.84ITNALYY191 pKa = 10.7AFLLVEE197 pKa = 4.28MARR200 pKa = 11.84PEE202 pKa = 4.25TQQGARR208 pKa = 11.84ALAAMRR214 pKa = 11.84INGTGMTMVGLFTQAAKK231 pKa = 10.89NLGATPSDD239 pKa = 3.95LLEE242 pKa = 4.48DD243 pKa = 3.49LCMRR247 pKa = 11.84SLVDD251 pKa = 3.14SARR254 pKa = 11.84RR255 pKa = 11.84IVRR258 pKa = 11.84LMVQVSQAEE267 pKa = 4.57TIQARR272 pKa = 11.84YY273 pKa = 9.15AVMMSRR279 pKa = 11.84MLNEE283 pKa = 4.63NYY285 pKa = 9.88FKK287 pKa = 10.9AYY289 pKa = 9.95GLNDD293 pKa = 3.3NSRR296 pKa = 11.84ISAILVAVNGHH307 pKa = 5.72FSEE310 pKa = 4.47DD311 pKa = 3.45TIEE314 pKa = 4.05ALEE317 pKa = 4.66GIKK320 pKa = 10.6VSAEE324 pKa = 3.8FADD327 pKa = 4.08LARR330 pKa = 11.84RR331 pKa = 11.84IAIALIEE338 pKa = 4.55KK339 pKa = 9.91YY340 pKa = 10.85DD341 pKa = 3.68NASDD345 pKa = 3.76GGEE348 pKa = 4.04GASEE352 pKa = 4.23IIKK355 pKa = 10.32SAVRR359 pKa = 11.84GSSGAFKK366 pKa = 10.72GSRR369 pKa = 11.84GKK371 pKa = 9.6PQGRR375 pKa = 11.84RR376 pKa = 11.84QEE378 pKa = 4.0ASGEE382 pKa = 4.01GDD384 pKa = 4.46LDD386 pKa = 4.99SEE388 pKa = 5.61DD389 pKa = 6.32DD390 pKa = 4.69DD391 pKa = 4.84DD392 pKa = 5.33QDD394 pKa = 3.64YY395 pKa = 11.68SKK397 pKa = 11.44YY398 pKa = 11.04AA399 pKa = 3.36
MM1 pKa = 7.77AFQKK5 pKa = 10.68EE6 pKa = 4.4FFGLRR11 pKa = 11.84DD12 pKa = 3.59VKK14 pKa = 11.12VDD16 pKa = 3.78LSAGEE21 pKa = 4.52GLDD24 pKa = 3.67FDD26 pKa = 4.51PSEE29 pKa = 4.52VEE31 pKa = 3.73LTVYY35 pKa = 9.26RR36 pKa = 11.84TGADD40 pKa = 3.17TDD42 pKa = 3.77GNTIIKK48 pKa = 10.2ALAAVGGPKK57 pKa = 9.72TNEE60 pKa = 3.79ALSVLLAFVTLGTDD74 pKa = 2.85QDD76 pKa = 3.88EE77 pKa = 4.66YY78 pKa = 9.81EE79 pKa = 4.19TRR81 pKa = 11.84IKK83 pKa = 10.12ILKK86 pKa = 9.84EE87 pKa = 3.14IGFSVKK93 pKa = 9.34EE94 pKa = 3.77VPMAKK99 pKa = 9.87DD100 pKa = 3.06ASSGIEE106 pKa = 3.71APLEE110 pKa = 3.88NVAALVKK117 pKa = 10.53PEE119 pKa = 3.92TVYY122 pKa = 10.85QVIRR126 pKa = 11.84GVLYY130 pKa = 9.55TCALFVKK137 pKa = 10.28YY138 pKa = 10.15NVEE141 pKa = 4.1KK142 pKa = 8.09MQKK145 pKa = 9.45YY146 pKa = 9.75IKK148 pKa = 10.5NKK150 pKa = 10.08LPALATSYY158 pKa = 10.53GVPEE162 pKa = 4.58LEE164 pKa = 4.23EE165 pKa = 4.53FPTEE169 pKa = 3.89SSALKK174 pKa = 10.48KK175 pKa = 10.17LASCIRR181 pKa = 11.84PGQRR185 pKa = 11.84ITNALYY191 pKa = 10.7AFLLVEE197 pKa = 4.28MARR200 pKa = 11.84PEE202 pKa = 4.25TQQGARR208 pKa = 11.84ALAAMRR214 pKa = 11.84INGTGMTMVGLFTQAAKK231 pKa = 10.89NLGATPSDD239 pKa = 3.95LLEE242 pKa = 4.48DD243 pKa = 3.49LCMRR247 pKa = 11.84SLVDD251 pKa = 3.14SARR254 pKa = 11.84RR255 pKa = 11.84IVRR258 pKa = 11.84LMVQVSQAEE267 pKa = 4.57TIQARR272 pKa = 11.84YY273 pKa = 9.15AVMMSRR279 pKa = 11.84MLNEE283 pKa = 4.63NYY285 pKa = 9.88FKK287 pKa = 10.9AYY289 pKa = 9.95GLNDD293 pKa = 3.3NSRR296 pKa = 11.84ISAILVAVNGHH307 pKa = 5.72FSEE310 pKa = 4.47DD311 pKa = 3.45TIEE314 pKa = 4.05ALEE317 pKa = 4.66GIKK320 pKa = 10.6VSAEE324 pKa = 3.8FADD327 pKa = 4.08LARR330 pKa = 11.84RR331 pKa = 11.84IAIALIEE338 pKa = 4.55KK339 pKa = 9.91YY340 pKa = 10.85DD341 pKa = 3.68NASDD345 pKa = 3.76GGEE348 pKa = 4.04GASEE352 pKa = 4.23IIKK355 pKa = 10.32SAVRR359 pKa = 11.84GSSGAFKK366 pKa = 10.72GSRR369 pKa = 11.84GKK371 pKa = 9.6PQGRR375 pKa = 11.84RR376 pKa = 11.84QEE378 pKa = 4.0ASGEE382 pKa = 4.01GDD384 pKa = 4.46LDD386 pKa = 4.99SEE388 pKa = 5.61DD389 pKa = 6.32DD390 pKa = 4.69DD391 pKa = 4.84DD392 pKa = 5.33QDD394 pKa = 3.64YY395 pKa = 11.68SKK397 pKa = 11.44YY398 pKa = 11.04AA399 pKa = 3.36
Molecular weight: 43.37 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q9QJT8|PHOSP_SHRV Phosphoprotein OS=Snakehead rhabdovirus OX=103603 GN=P PE=3 SV=1
MM1 pKa = 7.62PFIRR5 pKa = 11.84RR6 pKa = 11.84PKK8 pKa = 9.64RR9 pKa = 11.84AILIPPLHH17 pKa = 6.52LTLKK21 pKa = 10.75DD22 pKa = 3.55DD23 pKa = 4.57DD24 pKa = 4.1KK25 pKa = 11.56VLVVEE30 pKa = 4.56TVGTLIISGMTPSNLTEE47 pKa = 4.1KK48 pKa = 10.74LGLAMKK54 pKa = 9.94LASAILGGDD63 pKa = 3.32SHH65 pKa = 7.28PAFNPLVEE73 pKa = 4.37IFSGAMEE80 pKa = 4.72FGASVEE86 pKa = 4.39KK87 pKa = 10.73LDD89 pKa = 4.89FMTRR93 pKa = 11.84EE94 pKa = 3.98NKK96 pKa = 10.06VITTYY101 pKa = 10.88KK102 pKa = 8.52VARR105 pKa = 11.84GKK107 pKa = 10.31AVALSNFPMEE117 pKa = 4.17KK118 pKa = 9.55RR119 pKa = 11.84VGEE122 pKa = 4.07KK123 pKa = 10.34SYY125 pKa = 8.19TTQIRR130 pKa = 11.84NGSITYY136 pKa = 8.64TGSFLFSAEE145 pKa = 4.04HH146 pKa = 6.7VGLKK150 pKa = 10.46DD151 pKa = 3.55NRR153 pKa = 11.84SLFAAGEE160 pKa = 4.26GLRR163 pKa = 11.84EE164 pKa = 4.19SPDD167 pKa = 3.08MAQARR172 pKa = 11.84EE173 pKa = 4.15AFAGSLPKK181 pKa = 10.64GKK183 pKa = 10.07NEE185 pKa = 4.17SSRR188 pKa = 11.84KK189 pKa = 9.17
MM1 pKa = 7.62PFIRR5 pKa = 11.84RR6 pKa = 11.84PKK8 pKa = 9.64RR9 pKa = 11.84AILIPPLHH17 pKa = 6.52LTLKK21 pKa = 10.75DD22 pKa = 3.55DD23 pKa = 4.57DD24 pKa = 4.1KK25 pKa = 11.56VLVVEE30 pKa = 4.56TVGTLIISGMTPSNLTEE47 pKa = 4.1KK48 pKa = 10.74LGLAMKK54 pKa = 9.94LASAILGGDD63 pKa = 3.32SHH65 pKa = 7.28PAFNPLVEE73 pKa = 4.37IFSGAMEE80 pKa = 4.72FGASVEE86 pKa = 4.39KK87 pKa = 10.73LDD89 pKa = 4.89FMTRR93 pKa = 11.84EE94 pKa = 3.98NKK96 pKa = 10.06VITTYY101 pKa = 10.88KK102 pKa = 8.52VARR105 pKa = 11.84GKK107 pKa = 10.31AVALSNFPMEE117 pKa = 4.17KK118 pKa = 9.55RR119 pKa = 11.84VGEE122 pKa = 4.07KK123 pKa = 10.34SYY125 pKa = 8.19TTQIRR130 pKa = 11.84NGSITYY136 pKa = 8.64TGSFLFSAEE145 pKa = 4.04HH146 pKa = 6.7VGLKK150 pKa = 10.46DD151 pKa = 3.55NRR153 pKa = 11.84SLFAAGEE160 pKa = 4.26GLRR163 pKa = 11.84EE164 pKa = 4.19SPDD167 pKa = 3.08MAQARR172 pKa = 11.84EE173 pKa = 4.15AFAGSLPKK181 pKa = 10.64GKK183 pKa = 10.07NEE185 pKa = 4.17SSRR188 pKa = 11.84KK189 pKa = 9.17
Molecular weight: 20.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3425 |
122 |
1983 |
570.8 |
64.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.511 ± 1.086 | 1.547 ± 0.353 |
5.343 ± 0.407 | 7.504 ± 0.561 |
3.474 ± 0.348 | 6.19 ± 0.51 |
2.277 ± 0.406 | 6.102 ± 0.206 |
6.657 ± 0.549 | 10.891 ± 1.35 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.248 ± 0.408 | 3.533 ± 0.286 |
4.847 ± 0.753 | 3.299 ± 0.288 |
6.102 ± 0.512 | 7.62 ± 0.472 |
6.102 ± 0.385 | 5.752 ± 0.39 |
1.314 ± 0.358 | 2.686 ± 0.386 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
