
Quezon virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Hantaviridae; Mammantavirinae; Mobatvirus; Quezon mobatvirus
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
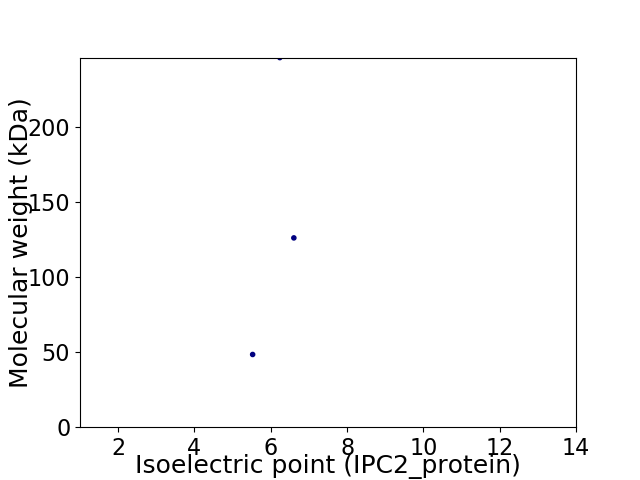
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B0WVN1|A0A1B0WVN1_9VIRU Envelopment polyprotein OS=Quezon virus OX=1841195 PE=3 SV=1
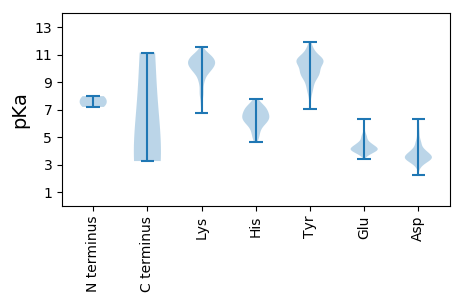
MM1 pKa = 7.58ASVKK5 pKa = 9.98EE6 pKa = 4.1LEE8 pKa = 4.45EE9 pKa = 3.78EE10 pKa = 3.89QQRR13 pKa = 11.84IEE15 pKa = 4.26RR16 pKa = 11.84EE17 pKa = 3.82LSIAQAKK24 pKa = 8.16LTDD27 pKa = 3.88AKK29 pKa = 11.16SKK31 pKa = 9.07ITEE34 pKa = 3.92SDD36 pKa = 3.38KK37 pKa = 11.4PDD39 pKa = 3.97DD40 pKa = 5.14LDD42 pKa = 3.53IKK44 pKa = 10.18MFNEE48 pKa = 3.81RR49 pKa = 11.84AEE51 pKa = 4.43TVSNLRR57 pKa = 11.84QRR59 pKa = 11.84LEE61 pKa = 3.72EE62 pKa = 4.19TKK64 pKa = 10.88RR65 pKa = 11.84MIAEE69 pKa = 4.78DD70 pKa = 3.12IKK72 pKa = 11.24KK73 pKa = 10.35SVNQKK78 pKa = 7.8PQKK81 pKa = 10.4KK82 pKa = 8.03GTDD85 pKa = 3.55LDD87 pKa = 4.3DD88 pKa = 5.96DD89 pKa = 3.84EE90 pKa = 6.32HH91 pKa = 5.75LTEE94 pKa = 4.96RR95 pKa = 11.84SSLRR99 pKa = 11.84YY100 pKa = 10.39GNVIDD105 pKa = 6.32LNDD108 pKa = 3.65TDD110 pKa = 3.93IDD112 pKa = 4.1EE113 pKa = 5.12PSGSSADD120 pKa = 3.51WISIVGYY127 pKa = 9.74LLSFSLVIILKK138 pKa = 9.85ALYY141 pKa = 9.36MLTTRR146 pKa = 11.84GRR148 pKa = 11.84QTIKK152 pKa = 10.1EE153 pKa = 4.12NKK155 pKa = 8.49GNRR158 pKa = 11.84IRR160 pKa = 11.84FKK162 pKa = 10.67DD163 pKa = 3.52DD164 pKa = 3.15SSFTEE169 pKa = 3.82KK170 pKa = 10.92GGVKK174 pKa = 9.43IPKK177 pKa = 8.92HH178 pKa = 6.18LYY180 pKa = 9.04VSLPTGQSSMKK191 pKa = 10.18ADD193 pKa = 4.65EE194 pKa = 4.32ITPGRR199 pKa = 11.84YY200 pKa = 7.53KK201 pKa = 10.36TAVCGLYY208 pKa = 9.78PAEE211 pKa = 4.18AKK213 pKa = 9.88ARR215 pKa = 11.84KK216 pKa = 8.8LVSPVMGVIGFNALVDD232 pKa = 3.45KK233 pKa = 11.12WEE235 pKa = 4.38DD236 pKa = 3.68TVSEE240 pKa = 4.39MMLEE244 pKa = 4.17PCQFIEE250 pKa = 4.74GNLMSQPEE258 pKa = 4.63DD259 pKa = 3.56LTNAAYY265 pKa = 9.18FAKK268 pKa = 10.41RR269 pKa = 11.84EE270 pKa = 3.81EE271 pKa = 4.31DD272 pKa = 3.35LRR274 pKa = 11.84GVRR277 pKa = 11.84SPDD280 pKa = 3.01MVQLYY285 pKa = 10.6DD286 pKa = 3.6LAKK289 pKa = 10.42SAGCKK294 pKa = 9.39LVNDD298 pKa = 3.74ITEE301 pKa = 4.12PHH303 pKa = 6.57APWVFACAPDD313 pKa = 3.87RR314 pKa = 11.84CPPTALYY321 pKa = 10.59VAGLAEE327 pKa = 4.62LGAFFSILQDD337 pKa = 3.28MRR339 pKa = 11.84NTIMASKK346 pKa = 10.53LVGTAEE352 pKa = 4.18EE353 pKa = 4.06KK354 pKa = 10.77LKK356 pKa = 10.81RR357 pKa = 11.84KK358 pKa = 9.86SSFYY362 pKa = 10.32QSYY365 pKa = 10.61LRR367 pKa = 11.84RR368 pKa = 11.84TQSMGIQLDD377 pKa = 3.23QRR379 pKa = 11.84IIVIFMLSWGKK390 pKa = 9.91MMTDD394 pKa = 3.66HH395 pKa = 6.84FHH397 pKa = 7.53LGDD400 pKa = 5.74DD401 pKa = 3.57MDD403 pKa = 6.0ADD405 pKa = 4.04LRR407 pKa = 11.84KK408 pKa = 9.93VCQSLIDD415 pKa = 3.96EE416 pKa = 4.49KK417 pKa = 11.32VKK419 pKa = 10.87QISNQDD425 pKa = 3.33ALKK428 pKa = 10.82LL429 pKa = 3.72
MM1 pKa = 7.58ASVKK5 pKa = 9.98EE6 pKa = 4.1LEE8 pKa = 4.45EE9 pKa = 3.78EE10 pKa = 3.89QQRR13 pKa = 11.84IEE15 pKa = 4.26RR16 pKa = 11.84EE17 pKa = 3.82LSIAQAKK24 pKa = 8.16LTDD27 pKa = 3.88AKK29 pKa = 11.16SKK31 pKa = 9.07ITEE34 pKa = 3.92SDD36 pKa = 3.38KK37 pKa = 11.4PDD39 pKa = 3.97DD40 pKa = 5.14LDD42 pKa = 3.53IKK44 pKa = 10.18MFNEE48 pKa = 3.81RR49 pKa = 11.84AEE51 pKa = 4.43TVSNLRR57 pKa = 11.84QRR59 pKa = 11.84LEE61 pKa = 3.72EE62 pKa = 4.19TKK64 pKa = 10.88RR65 pKa = 11.84MIAEE69 pKa = 4.78DD70 pKa = 3.12IKK72 pKa = 11.24KK73 pKa = 10.35SVNQKK78 pKa = 7.8PQKK81 pKa = 10.4KK82 pKa = 8.03GTDD85 pKa = 3.55LDD87 pKa = 4.3DD88 pKa = 5.96DD89 pKa = 3.84EE90 pKa = 6.32HH91 pKa = 5.75LTEE94 pKa = 4.96RR95 pKa = 11.84SSLRR99 pKa = 11.84YY100 pKa = 10.39GNVIDD105 pKa = 6.32LNDD108 pKa = 3.65TDD110 pKa = 3.93IDD112 pKa = 4.1EE113 pKa = 5.12PSGSSADD120 pKa = 3.51WISIVGYY127 pKa = 9.74LLSFSLVIILKK138 pKa = 9.85ALYY141 pKa = 9.36MLTTRR146 pKa = 11.84GRR148 pKa = 11.84QTIKK152 pKa = 10.1EE153 pKa = 4.12NKK155 pKa = 8.49GNRR158 pKa = 11.84IRR160 pKa = 11.84FKK162 pKa = 10.67DD163 pKa = 3.52DD164 pKa = 3.15SSFTEE169 pKa = 3.82KK170 pKa = 10.92GGVKK174 pKa = 9.43IPKK177 pKa = 8.92HH178 pKa = 6.18LYY180 pKa = 9.04VSLPTGQSSMKK191 pKa = 10.18ADD193 pKa = 4.65EE194 pKa = 4.32ITPGRR199 pKa = 11.84YY200 pKa = 7.53KK201 pKa = 10.36TAVCGLYY208 pKa = 9.78PAEE211 pKa = 4.18AKK213 pKa = 9.88ARR215 pKa = 11.84KK216 pKa = 8.8LVSPVMGVIGFNALVDD232 pKa = 3.45KK233 pKa = 11.12WEE235 pKa = 4.38DD236 pKa = 3.68TVSEE240 pKa = 4.39MMLEE244 pKa = 4.17PCQFIEE250 pKa = 4.74GNLMSQPEE258 pKa = 4.63DD259 pKa = 3.56LTNAAYY265 pKa = 9.18FAKK268 pKa = 10.41RR269 pKa = 11.84EE270 pKa = 3.81EE271 pKa = 4.31DD272 pKa = 3.35LRR274 pKa = 11.84GVRR277 pKa = 11.84SPDD280 pKa = 3.01MVQLYY285 pKa = 10.6DD286 pKa = 3.6LAKK289 pKa = 10.42SAGCKK294 pKa = 9.39LVNDD298 pKa = 3.74ITEE301 pKa = 4.12PHH303 pKa = 6.57APWVFACAPDD313 pKa = 3.87RR314 pKa = 11.84CPPTALYY321 pKa = 10.59VAGLAEE327 pKa = 4.62LGAFFSILQDD337 pKa = 3.28MRR339 pKa = 11.84NTIMASKK346 pKa = 10.53LVGTAEE352 pKa = 4.18EE353 pKa = 4.06KK354 pKa = 10.77LKK356 pKa = 10.81RR357 pKa = 11.84KK358 pKa = 9.86SSFYY362 pKa = 10.32QSYY365 pKa = 10.61LRR367 pKa = 11.84RR368 pKa = 11.84TQSMGIQLDD377 pKa = 3.23QRR379 pKa = 11.84IIVIFMLSWGKK390 pKa = 9.91MMTDD394 pKa = 3.66HH395 pKa = 6.84FHH397 pKa = 7.53LGDD400 pKa = 5.74DD401 pKa = 3.57MDD403 pKa = 6.0ADD405 pKa = 4.04LRR407 pKa = 11.84KK408 pKa = 9.93VCQSLIDD415 pKa = 3.96EE416 pKa = 4.49KK417 pKa = 11.32VKK419 pKa = 10.87QISNQDD425 pKa = 3.33ALKK428 pKa = 10.82LL429 pKa = 3.72
Molecular weight: 48.44 kDa
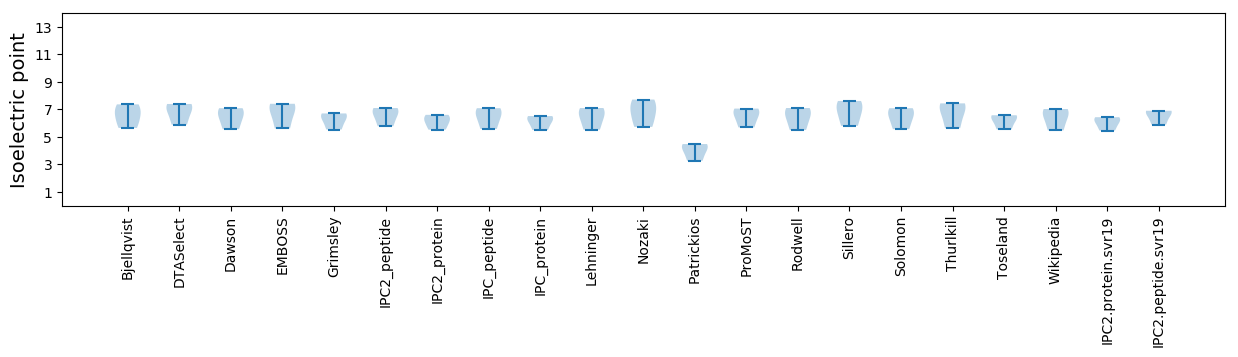
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B0WVN3|A0A1B0WVN3_9VIRU Replicase OS=Quezon virus OX=1841195 PE=4 SV=1
MM1 pKa = 7.97RR2 pKa = 11.84LILWMLILGFTDD14 pKa = 3.3QKK16 pKa = 11.42SLLEE20 pKa = 4.12IKK22 pKa = 7.84FTCPHH27 pKa = 6.33DD28 pKa = 3.64TRR30 pKa = 11.84FEE32 pKa = 4.13TGFGYY37 pKa = 10.7GAVDD41 pKa = 4.12LPSITEE47 pKa = 3.92ASDD50 pKa = 4.47LIVPVEE56 pKa = 4.16TSCPFEE62 pKa = 4.16FNLVRR67 pKa = 11.84KK68 pKa = 9.21QVTSVKK74 pKa = 9.5MKK76 pKa = 7.34WWEE79 pKa = 4.06RR80 pKa = 11.84KK81 pKa = 9.5SSVAAGTNADD91 pKa = 3.37QNTYY95 pKa = 8.2EE96 pKa = 4.61EE97 pKa = 4.5KK98 pKa = 10.73EE99 pKa = 4.03KK100 pKa = 11.12DD101 pKa = 2.97AGAIGICNIQHH112 pKa = 6.59NDD114 pKa = 2.7ITSISQNRR122 pKa = 11.84RR123 pKa = 11.84GLMCLEE129 pKa = 4.11YY130 pKa = 10.73SCNLTHH136 pKa = 7.29CMPALHH142 pKa = 6.64VIIPHH147 pKa = 5.59QICMNIRR154 pKa = 11.84TCLLTWKK161 pKa = 8.59STKK164 pKa = 9.44IDD166 pKa = 4.5LIFEE170 pKa = 5.1RR171 pKa = 11.84IFCPHH176 pKa = 7.37GILVAGNCFQPYY188 pKa = 9.93YY189 pKa = 10.61GVTSPPPEE197 pKa = 4.01SEE199 pKa = 3.96RR200 pKa = 11.84AEE202 pKa = 4.22LSTTCFLITTKK213 pKa = 10.08QSKK216 pKa = 10.28AEE218 pKa = 4.05KK219 pKa = 10.03NLPQLVTAFQSEE231 pKa = 4.48ITGNTCSPVNMAGYY245 pKa = 8.82YY246 pKa = 9.31SCFIKK251 pKa = 10.54GYY253 pKa = 8.52SQMINVPGHH262 pKa = 6.5KK263 pKa = 10.04DD264 pKa = 3.26AITGEE269 pKa = 4.11IMSKK273 pKa = 10.0IIKK276 pKa = 9.7FSHH279 pKa = 6.6GEE281 pKa = 3.73DD282 pKa = 3.17HH283 pKa = 7.75DD284 pKa = 5.31RR285 pKa = 11.84YY286 pKa = 8.72TDD288 pKa = 3.16GHH290 pKa = 6.79GILNVAGPLNFSLKK304 pKa = 9.75STDD307 pKa = 3.26KK308 pKa = 10.97FSGVCYY314 pKa = 10.52SGDD317 pKa = 3.16ILYY320 pKa = 9.64TSLYY324 pKa = 9.68MNTKK328 pKa = 10.23KK329 pKa = 10.81SGDD332 pKa = 3.74DD333 pKa = 3.54FTVVLAKK340 pKa = 10.71GIIPNTDD347 pKa = 2.81HH348 pKa = 7.19SSCDD352 pKa = 3.5MKK354 pKa = 11.12LLPLVWQGMINIPGYY369 pKa = 8.89IEE371 pKa = 4.6KK372 pKa = 9.76MEE374 pKa = 4.37PCKK377 pKa = 10.77VFCTLSGPGASCEE390 pKa = 4.26AFSSTGIFNISSDD403 pKa = 3.18TCLIGKK409 pKa = 7.18LHH411 pKa = 7.03RR412 pKa = 11.84YY413 pKa = 8.4KK414 pKa = 10.68HH415 pKa = 6.44LEE417 pKa = 3.87DD418 pKa = 4.01QISFICQRR426 pKa = 11.84VDD428 pKa = 2.91QDD430 pKa = 4.52LVIYY434 pKa = 10.72CNGQKK439 pKa = 10.31KK440 pKa = 9.91VIKK443 pKa = 9.84TNTLVIGQCIYY454 pKa = 10.59TITSALSLLPTIAHH468 pKa = 6.88PLAVEE473 pKa = 4.12LCVTGFHH480 pKa = 6.43GWATLLLLITFCFGWLLIPLVSYY503 pKa = 9.85LVIQVLRR510 pKa = 11.84LILILMNKK518 pKa = 8.22HH519 pKa = 5.44TGQSRR524 pKa = 11.84FEE526 pKa = 4.64NILKK530 pKa = 10.21KK531 pKa = 10.91LKK533 pKa = 10.72DD534 pKa = 3.87EE535 pKa = 4.49FQHH538 pKa = 6.43TIGSTTCSYY547 pKa = 11.26CKK549 pKa = 9.72TDD551 pKa = 3.38CCSKK555 pKa = 11.22EE556 pKa = 3.81EE557 pKa = 4.16LTSHH561 pKa = 6.88EE562 pKa = 4.59EE563 pKa = 3.84NCSKK567 pKa = 10.88GKK569 pKa = 10.38CPYY572 pKa = 9.2CLKK575 pKa = 10.49EE576 pKa = 4.27IGSSSIIATEE586 pKa = 3.89HH587 pKa = 6.48FKK589 pKa = 10.31ICPLIDD595 pKa = 3.41RR596 pKa = 11.84FITKK600 pKa = 9.6IRR602 pKa = 11.84QNVASTPTHH611 pKa = 5.12GTLLYY616 pKa = 10.3RR617 pKa = 11.84KK618 pKa = 9.64LGTFRR623 pKa = 11.84YY624 pKa = 9.73KK625 pKa = 10.39NRR627 pKa = 11.84CFIFTIWTVLIIIEE641 pKa = 4.28SLVWAVSAQKK651 pKa = 8.3MTVDD655 pKa = 4.58PGWSDD660 pKa = 3.27TAHH663 pKa = 6.4GVGSVPMEE671 pKa = 4.44LDD673 pKa = 3.46YY674 pKa = 11.49EE675 pKa = 4.51LDD677 pKa = 3.5FALVSGSTFIHH688 pKa = 6.42KK689 pKa = 10.3RR690 pKa = 11.84LLQSPQAKK698 pKa = 9.1EE699 pKa = 3.48HH700 pKa = 6.43TIPFTVTIDD709 pKa = 3.61TQKK712 pKa = 9.06ITAVVQTLGHH722 pKa = 6.04WMDD725 pKa = 4.09AEE727 pKa = 4.51VNLKK731 pKa = 10.45SVFHH735 pKa = 7.12CYY737 pKa = 9.53GACKK741 pKa = 9.77AYY743 pKa = 9.79KK744 pKa = 10.26YY745 pKa = 9.08PWQEE749 pKa = 3.92AYY751 pKa = 10.33CSHH754 pKa = 6.05EE755 pKa = 3.63VDD757 pKa = 4.58YY758 pKa = 10.55EE759 pKa = 4.32YY760 pKa = 11.15QSSWSCNPPTCPGISTGCTACGMYY784 pKa = 10.24LDD786 pKa = 4.7KK787 pKa = 10.4LTPKK791 pKa = 10.5ASVYY795 pKa = 10.55KK796 pKa = 10.29IINIKK801 pKa = 8.17YY802 pKa = 9.64SRR804 pKa = 11.84YY805 pKa = 9.12ICYY808 pKa = 10.05QLGTEE813 pKa = 4.31KK814 pKa = 10.34QCKK817 pKa = 7.9EE818 pKa = 4.05VEE820 pKa = 4.3GNDD823 pKa = 3.73CVTGKK828 pKa = 9.79HH829 pKa = 6.03FKK831 pKa = 10.22ACLVGTVSNIVASDD845 pKa = 3.46TLVFLGPLDD854 pKa = 4.16AGALIMKK861 pKa = 7.33QWCVSNCKK869 pKa = 10.37LGDD872 pKa = 3.93PGDD875 pKa = 3.69IMSVQDD881 pKa = 5.01KK882 pKa = 10.26INCPSYY888 pKa = 10.16EE889 pKa = 3.85GTINRR894 pKa = 11.84VCRR897 pKa = 11.84FATMPVCSYY906 pKa = 10.74QGNQVPGFKK915 pKa = 10.69KK916 pKa = 10.1MMATKK921 pKa = 10.55DD922 pKa = 3.38SFLSVNMSNVKK933 pKa = 10.45LSLSTMSWEE942 pKa = 4.94DD943 pKa = 3.19PDD945 pKa = 5.81SIYY948 pKa = 10.62KK949 pKa = 10.08DD950 pKa = 3.54HH951 pKa = 7.21INIVVTKK958 pKa = 10.47DD959 pKa = 3.13LDD961 pKa = 3.85FEE963 pKa = 4.39EE964 pKa = 5.93LSEE967 pKa = 4.31NPCSVEE973 pKa = 4.1SVISSIEE980 pKa = 4.03GSWGSGVGFTITCSISLTEE999 pKa = 4.71CSDD1002 pKa = 3.53FLTTIKK1008 pKa = 10.86ACDD1011 pKa = 3.44DD1012 pKa = 4.57AICYY1016 pKa = 9.23GGKK1019 pKa = 9.86AVRR1022 pKa = 11.84LLRR1025 pKa = 11.84GQNTVKK1031 pKa = 9.78VQGRR1035 pKa = 11.84GGHH1038 pKa = 5.75SGSKK1042 pKa = 9.81FRR1044 pKa = 11.84CCHH1047 pKa = 5.97EE1048 pKa = 4.22NHH1050 pKa = 6.9CSTKK1054 pKa = 10.32GLRR1057 pKa = 11.84ASSPHH1062 pKa = 6.62LEE1064 pKa = 4.38RR1065 pKa = 11.84IGDD1068 pKa = 3.69EE1069 pKa = 4.06RR1070 pKa = 11.84ALEE1073 pKa = 4.13SQVFSDD1079 pKa = 4.87GAPEE1083 pKa = 4.61CGISCQFKK1091 pKa = 10.74KK1092 pKa = 10.1VWEE1095 pKa = 4.18WLTGIFSGNWFILIVLVIVMIISIIILSFFCPAKK1129 pKa = 10.44RR1130 pKa = 11.84KK1131 pKa = 8.02TSS1133 pKa = 3.25
MM1 pKa = 7.97RR2 pKa = 11.84LILWMLILGFTDD14 pKa = 3.3QKK16 pKa = 11.42SLLEE20 pKa = 4.12IKK22 pKa = 7.84FTCPHH27 pKa = 6.33DD28 pKa = 3.64TRR30 pKa = 11.84FEE32 pKa = 4.13TGFGYY37 pKa = 10.7GAVDD41 pKa = 4.12LPSITEE47 pKa = 3.92ASDD50 pKa = 4.47LIVPVEE56 pKa = 4.16TSCPFEE62 pKa = 4.16FNLVRR67 pKa = 11.84KK68 pKa = 9.21QVTSVKK74 pKa = 9.5MKK76 pKa = 7.34WWEE79 pKa = 4.06RR80 pKa = 11.84KK81 pKa = 9.5SSVAAGTNADD91 pKa = 3.37QNTYY95 pKa = 8.2EE96 pKa = 4.61EE97 pKa = 4.5KK98 pKa = 10.73EE99 pKa = 4.03KK100 pKa = 11.12DD101 pKa = 2.97AGAIGICNIQHH112 pKa = 6.59NDD114 pKa = 2.7ITSISQNRR122 pKa = 11.84RR123 pKa = 11.84GLMCLEE129 pKa = 4.11YY130 pKa = 10.73SCNLTHH136 pKa = 7.29CMPALHH142 pKa = 6.64VIIPHH147 pKa = 5.59QICMNIRR154 pKa = 11.84TCLLTWKK161 pKa = 8.59STKK164 pKa = 9.44IDD166 pKa = 4.5LIFEE170 pKa = 5.1RR171 pKa = 11.84IFCPHH176 pKa = 7.37GILVAGNCFQPYY188 pKa = 9.93YY189 pKa = 10.61GVTSPPPEE197 pKa = 4.01SEE199 pKa = 3.96RR200 pKa = 11.84AEE202 pKa = 4.22LSTTCFLITTKK213 pKa = 10.08QSKK216 pKa = 10.28AEE218 pKa = 4.05KK219 pKa = 10.03NLPQLVTAFQSEE231 pKa = 4.48ITGNTCSPVNMAGYY245 pKa = 8.82YY246 pKa = 9.31SCFIKK251 pKa = 10.54GYY253 pKa = 8.52SQMINVPGHH262 pKa = 6.5KK263 pKa = 10.04DD264 pKa = 3.26AITGEE269 pKa = 4.11IMSKK273 pKa = 10.0IIKK276 pKa = 9.7FSHH279 pKa = 6.6GEE281 pKa = 3.73DD282 pKa = 3.17HH283 pKa = 7.75DD284 pKa = 5.31RR285 pKa = 11.84YY286 pKa = 8.72TDD288 pKa = 3.16GHH290 pKa = 6.79GILNVAGPLNFSLKK304 pKa = 9.75STDD307 pKa = 3.26KK308 pKa = 10.97FSGVCYY314 pKa = 10.52SGDD317 pKa = 3.16ILYY320 pKa = 9.64TSLYY324 pKa = 9.68MNTKK328 pKa = 10.23KK329 pKa = 10.81SGDD332 pKa = 3.74DD333 pKa = 3.54FTVVLAKK340 pKa = 10.71GIIPNTDD347 pKa = 2.81HH348 pKa = 7.19SSCDD352 pKa = 3.5MKK354 pKa = 11.12LLPLVWQGMINIPGYY369 pKa = 8.89IEE371 pKa = 4.6KK372 pKa = 9.76MEE374 pKa = 4.37PCKK377 pKa = 10.77VFCTLSGPGASCEE390 pKa = 4.26AFSSTGIFNISSDD403 pKa = 3.18TCLIGKK409 pKa = 7.18LHH411 pKa = 7.03RR412 pKa = 11.84YY413 pKa = 8.4KK414 pKa = 10.68HH415 pKa = 6.44LEE417 pKa = 3.87DD418 pKa = 4.01QISFICQRR426 pKa = 11.84VDD428 pKa = 2.91QDD430 pKa = 4.52LVIYY434 pKa = 10.72CNGQKK439 pKa = 10.31KK440 pKa = 9.91VIKK443 pKa = 9.84TNTLVIGQCIYY454 pKa = 10.59TITSALSLLPTIAHH468 pKa = 6.88PLAVEE473 pKa = 4.12LCVTGFHH480 pKa = 6.43GWATLLLLITFCFGWLLIPLVSYY503 pKa = 9.85LVIQVLRR510 pKa = 11.84LILILMNKK518 pKa = 8.22HH519 pKa = 5.44TGQSRR524 pKa = 11.84FEE526 pKa = 4.64NILKK530 pKa = 10.21KK531 pKa = 10.91LKK533 pKa = 10.72DD534 pKa = 3.87EE535 pKa = 4.49FQHH538 pKa = 6.43TIGSTTCSYY547 pKa = 11.26CKK549 pKa = 9.72TDD551 pKa = 3.38CCSKK555 pKa = 11.22EE556 pKa = 3.81EE557 pKa = 4.16LTSHH561 pKa = 6.88EE562 pKa = 4.59EE563 pKa = 3.84NCSKK567 pKa = 10.88GKK569 pKa = 10.38CPYY572 pKa = 9.2CLKK575 pKa = 10.49EE576 pKa = 4.27IGSSSIIATEE586 pKa = 3.89HH587 pKa = 6.48FKK589 pKa = 10.31ICPLIDD595 pKa = 3.41RR596 pKa = 11.84FITKK600 pKa = 9.6IRR602 pKa = 11.84QNVASTPTHH611 pKa = 5.12GTLLYY616 pKa = 10.3RR617 pKa = 11.84KK618 pKa = 9.64LGTFRR623 pKa = 11.84YY624 pKa = 9.73KK625 pKa = 10.39NRR627 pKa = 11.84CFIFTIWTVLIIIEE641 pKa = 4.28SLVWAVSAQKK651 pKa = 8.3MTVDD655 pKa = 4.58PGWSDD660 pKa = 3.27TAHH663 pKa = 6.4GVGSVPMEE671 pKa = 4.44LDD673 pKa = 3.46YY674 pKa = 11.49EE675 pKa = 4.51LDD677 pKa = 3.5FALVSGSTFIHH688 pKa = 6.42KK689 pKa = 10.3RR690 pKa = 11.84LLQSPQAKK698 pKa = 9.1EE699 pKa = 3.48HH700 pKa = 6.43TIPFTVTIDD709 pKa = 3.61TQKK712 pKa = 9.06ITAVVQTLGHH722 pKa = 6.04WMDD725 pKa = 4.09AEE727 pKa = 4.51VNLKK731 pKa = 10.45SVFHH735 pKa = 7.12CYY737 pKa = 9.53GACKK741 pKa = 9.77AYY743 pKa = 9.79KK744 pKa = 10.26YY745 pKa = 9.08PWQEE749 pKa = 3.92AYY751 pKa = 10.33CSHH754 pKa = 6.05EE755 pKa = 3.63VDD757 pKa = 4.58YY758 pKa = 10.55EE759 pKa = 4.32YY760 pKa = 11.15QSSWSCNPPTCPGISTGCTACGMYY784 pKa = 10.24LDD786 pKa = 4.7KK787 pKa = 10.4LTPKK791 pKa = 10.5ASVYY795 pKa = 10.55KK796 pKa = 10.29IINIKK801 pKa = 8.17YY802 pKa = 9.64SRR804 pKa = 11.84YY805 pKa = 9.12ICYY808 pKa = 10.05QLGTEE813 pKa = 4.31KK814 pKa = 10.34QCKK817 pKa = 7.9EE818 pKa = 4.05VEE820 pKa = 4.3GNDD823 pKa = 3.73CVTGKK828 pKa = 9.79HH829 pKa = 6.03FKK831 pKa = 10.22ACLVGTVSNIVASDD845 pKa = 3.46TLVFLGPLDD854 pKa = 4.16AGALIMKK861 pKa = 7.33QWCVSNCKK869 pKa = 10.37LGDD872 pKa = 3.93PGDD875 pKa = 3.69IMSVQDD881 pKa = 5.01KK882 pKa = 10.26INCPSYY888 pKa = 10.16EE889 pKa = 3.85GTINRR894 pKa = 11.84VCRR897 pKa = 11.84FATMPVCSYY906 pKa = 10.74QGNQVPGFKK915 pKa = 10.69KK916 pKa = 10.1MMATKK921 pKa = 10.55DD922 pKa = 3.38SFLSVNMSNVKK933 pKa = 10.45LSLSTMSWEE942 pKa = 4.94DD943 pKa = 3.19PDD945 pKa = 5.81SIYY948 pKa = 10.62KK949 pKa = 10.08DD950 pKa = 3.54HH951 pKa = 7.21INIVVTKK958 pKa = 10.47DD959 pKa = 3.13LDD961 pKa = 3.85FEE963 pKa = 4.39EE964 pKa = 5.93LSEE967 pKa = 4.31NPCSVEE973 pKa = 4.1SVISSIEE980 pKa = 4.03GSWGSGVGFTITCSISLTEE999 pKa = 4.71CSDD1002 pKa = 3.53FLTTIKK1008 pKa = 10.86ACDD1011 pKa = 3.44DD1012 pKa = 4.57AICYY1016 pKa = 9.23GGKK1019 pKa = 9.86AVRR1022 pKa = 11.84LLRR1025 pKa = 11.84GQNTVKK1031 pKa = 9.78VQGRR1035 pKa = 11.84GGHH1038 pKa = 5.75SGSKK1042 pKa = 9.81FRR1044 pKa = 11.84CCHH1047 pKa = 5.97EE1048 pKa = 4.22NHH1050 pKa = 6.9CSTKK1054 pKa = 10.32GLRR1057 pKa = 11.84ASSPHH1062 pKa = 6.62LEE1064 pKa = 4.38RR1065 pKa = 11.84IGDD1068 pKa = 3.69EE1069 pKa = 4.06RR1070 pKa = 11.84ALEE1073 pKa = 4.13SQVFSDD1079 pKa = 4.87GAPEE1083 pKa = 4.61CGISCQFKK1091 pKa = 10.74KK1092 pKa = 10.1VWEE1095 pKa = 4.18WLTGIFSGNWFILIVLVIVMIISIIILSFFCPAKK1129 pKa = 10.44RR1130 pKa = 11.84KK1131 pKa = 8.02TSS1133 pKa = 3.25
Molecular weight: 126.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3709 |
429 |
2147 |
1236.3 |
140.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.311 ± 0.455 | 2.615 ± 1.039 |
5.797 ± 0.638 | 6.066 ± 0.443 |
4.691 ± 0.491 | 5.392 ± 0.601 |
2.453 ± 0.293 | 7.28 ± 0.493 |
7.468 ± 0.309 | 9.517 ± 0.451 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.723 ± 0.287 | 3.963 ± 0.29 |
3.667 ± 0.134 | 3.748 ± 0.183 |
3.99 ± 0.517 | 8.061 ± 0.261 |
5.851 ± 0.641 | 6.309 ± 0.283 |
1.456 ± 0.126 | 3.64 ± 0.358 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
