
Actinoalloteichus hoggarensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Pseudonocardiales; Pseudonocardiaceae; Actinoalloteichus
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
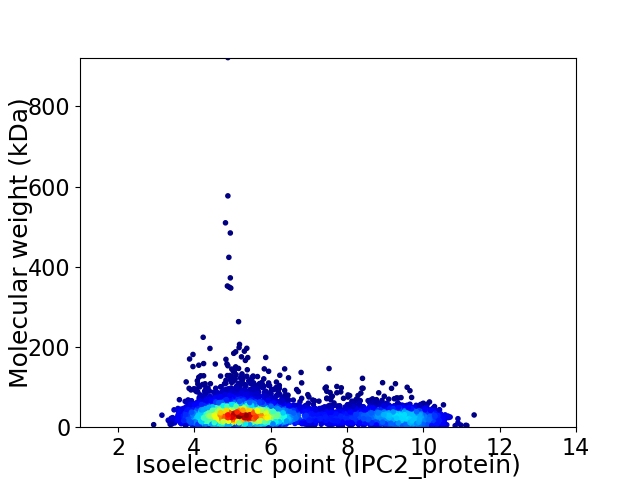
Virtual 2D-PAGE plot for 5520 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A221VX27|A0A221VX27_9PSEU UDP-glucose 4-epimerase OS=Actinoalloteichus hoggarensis OX=1470176 GN=AHOG_02225 PE=4 SV=1
MM1 pKa = 8.13RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84TLNAMSVMGLASALALTACGSGGGDD29 pKa = 3.29GDD31 pKa = 3.99GPAAVTFWDD40 pKa = 3.62TSGPAEE46 pKa = 4.11SPVFARR52 pKa = 11.84LAQDD56 pKa = 3.36CGEE59 pKa = 4.26AGGYY63 pKa = 8.82QVDD66 pKa = 3.81IEE68 pKa = 4.41QVAFDD73 pKa = 3.77QALNNYY79 pKa = 9.01RR80 pKa = 11.84NAAQGGQGPDD90 pKa = 2.92VFRR93 pKa = 11.84AEE95 pKa = 4.49VAWVPQLAEE104 pKa = 4.01NGLIQDD110 pKa = 4.69LSEE113 pKa = 4.36TDD115 pKa = 3.68LADD118 pKa = 3.84PADD121 pKa = 4.04FLEE124 pKa = 4.99TPWSSTQYY132 pKa = 10.87DD133 pKa = 3.3GRR135 pKa = 11.84TYY137 pKa = 10.92GVPQVTDD144 pKa = 3.57TLALLYY150 pKa = 10.93NRR152 pKa = 11.84DD153 pKa = 3.93LLDD156 pKa = 3.82EE157 pKa = 5.29AGVEE161 pKa = 4.56PPTSWAEE168 pKa = 4.07LAEE171 pKa = 4.31SAPPLGGEE179 pKa = 4.42SAIFLNNDD187 pKa = 2.72AYY189 pKa = 10.76YY190 pKa = 11.01ALPFVYY196 pKa = 9.92GAGGDD201 pKa = 3.93LVDD204 pKa = 4.99TEE206 pKa = 4.24AQEE209 pKa = 4.17ILVNSPEE216 pKa = 4.05AVEE219 pKa = 4.62GLTTAKK225 pKa = 10.7DD226 pKa = 3.76LLDD229 pKa = 3.95EE230 pKa = 4.91GAAQTALDD238 pKa = 3.69QGNSYY243 pKa = 11.63GNMKK247 pKa = 10.29AAFVAGDD254 pKa = 3.29VAMIIDD260 pKa = 4.54GPWAVAEE267 pKa = 4.38LLEE270 pKa = 5.45GDD272 pKa = 4.47AFADD276 pKa = 3.91PEE278 pKa = 4.32NLGIAPVPGPEE289 pKa = 4.37DD290 pKa = 3.4GAGNSPVGGHH300 pKa = 7.16DD301 pKa = 3.34YY302 pKa = 11.11VIRR305 pKa = 11.84QGSAATEE312 pKa = 4.1SAQDD316 pKa = 4.7LIACLSSTEE325 pKa = 4.07SQITVATEE333 pKa = 3.99LGLLPTRR340 pKa = 11.84EE341 pKa = 3.88SAYY344 pKa = 10.66ADD346 pKa = 3.64PAVAAQPIVSAFSPVIEE363 pKa = 4.42RR364 pKa = 11.84AHH366 pKa = 5.63PRR368 pKa = 11.84PWIPEE373 pKa = 3.93GGEE376 pKa = 3.84LFDD379 pKa = 4.83PLKK382 pKa = 10.48IAYY385 pKa = 9.95SDD387 pKa = 3.21ILAGNVEE394 pKa = 4.29PQDD397 pKa = 3.97ALDD400 pKa = 4.0EE401 pKa = 4.4VATIYY406 pKa = 10.73QDD408 pKa = 3.79TILTEE413 pKa = 4.27YY414 pKa = 10.71SVSS417 pKa = 3.38
MM1 pKa = 8.13RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84TLNAMSVMGLASALALTACGSGGGDD29 pKa = 3.29GDD31 pKa = 3.99GPAAVTFWDD40 pKa = 3.62TSGPAEE46 pKa = 4.11SPVFARR52 pKa = 11.84LAQDD56 pKa = 3.36CGEE59 pKa = 4.26AGGYY63 pKa = 8.82QVDD66 pKa = 3.81IEE68 pKa = 4.41QVAFDD73 pKa = 3.77QALNNYY79 pKa = 9.01RR80 pKa = 11.84NAAQGGQGPDD90 pKa = 2.92VFRR93 pKa = 11.84AEE95 pKa = 4.49VAWVPQLAEE104 pKa = 4.01NGLIQDD110 pKa = 4.69LSEE113 pKa = 4.36TDD115 pKa = 3.68LADD118 pKa = 3.84PADD121 pKa = 4.04FLEE124 pKa = 4.99TPWSSTQYY132 pKa = 10.87DD133 pKa = 3.3GRR135 pKa = 11.84TYY137 pKa = 10.92GVPQVTDD144 pKa = 3.57TLALLYY150 pKa = 10.93NRR152 pKa = 11.84DD153 pKa = 3.93LLDD156 pKa = 3.82EE157 pKa = 5.29AGVEE161 pKa = 4.56PPTSWAEE168 pKa = 4.07LAEE171 pKa = 4.31SAPPLGGEE179 pKa = 4.42SAIFLNNDD187 pKa = 2.72AYY189 pKa = 10.76YY190 pKa = 11.01ALPFVYY196 pKa = 9.92GAGGDD201 pKa = 3.93LVDD204 pKa = 4.99TEE206 pKa = 4.24AQEE209 pKa = 4.17ILVNSPEE216 pKa = 4.05AVEE219 pKa = 4.62GLTTAKK225 pKa = 10.7DD226 pKa = 3.76LLDD229 pKa = 3.95EE230 pKa = 4.91GAAQTALDD238 pKa = 3.69QGNSYY243 pKa = 11.63GNMKK247 pKa = 10.29AAFVAGDD254 pKa = 3.29VAMIIDD260 pKa = 4.54GPWAVAEE267 pKa = 4.38LLEE270 pKa = 5.45GDD272 pKa = 4.47AFADD276 pKa = 3.91PEE278 pKa = 4.32NLGIAPVPGPEE289 pKa = 4.37DD290 pKa = 3.4GAGNSPVGGHH300 pKa = 7.16DD301 pKa = 3.34YY302 pKa = 11.11VIRR305 pKa = 11.84QGSAATEE312 pKa = 4.1SAQDD316 pKa = 4.7LIACLSSTEE325 pKa = 4.07SQITVATEE333 pKa = 3.99LGLLPTRR340 pKa = 11.84EE341 pKa = 3.88SAYY344 pKa = 10.66ADD346 pKa = 3.64PAVAAQPIVSAFSPVIEE363 pKa = 4.42RR364 pKa = 11.84AHH366 pKa = 5.63PRR368 pKa = 11.84PWIPEE373 pKa = 3.93GGEE376 pKa = 3.84LFDD379 pKa = 4.83PLKK382 pKa = 10.48IAYY385 pKa = 9.95SDD387 pKa = 3.21ILAGNVEE394 pKa = 4.29PQDD397 pKa = 3.97ALDD400 pKa = 4.0EE401 pKa = 4.4VATIYY406 pKa = 10.73QDD408 pKa = 3.79TILTEE413 pKa = 4.27YY414 pKa = 10.71SVSS417 pKa = 3.38
Molecular weight: 43.59 kDa
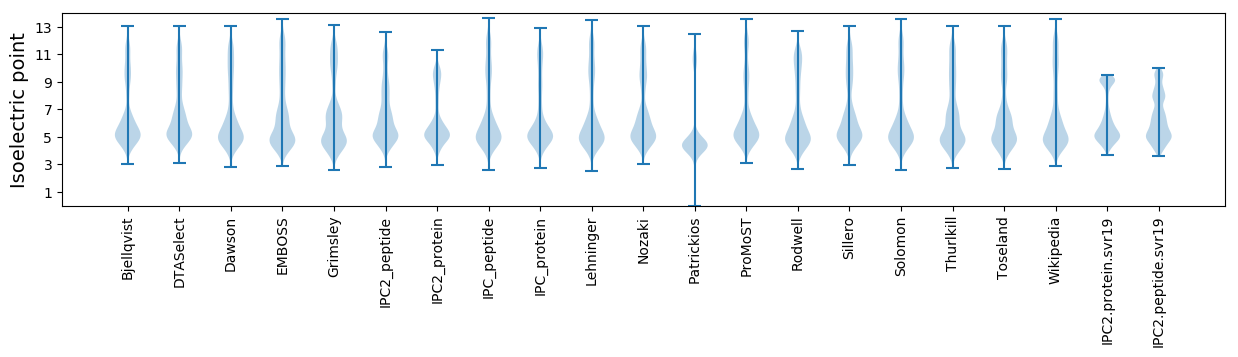
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A221VW50|A0A221VW50_9PSEU 3-oxoacyl-[acyl-carrier-protein] reductase FabG OS=Actinoalloteichus hoggarensis OX=1470176 GN=fabG1 PE=3 SV=1
MM1 pKa = 7.47SSRR4 pKa = 11.84RR5 pKa = 11.84SGTTTPPARR14 pKa = 11.84ALRR17 pKa = 11.84RR18 pKa = 11.84PRR20 pKa = 11.84TTSRR24 pKa = 11.84AGLRR28 pKa = 11.84RR29 pKa = 11.84VPTASPRR36 pKa = 11.84PVAGTLPAKK45 pKa = 10.39ARR47 pKa = 11.84TISVPAAQRR56 pKa = 11.84EE57 pKa = 4.45DD58 pKa = 2.99IRR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84APGRR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84ARR70 pKa = 11.84RR71 pKa = 11.84SRR73 pKa = 11.84RR74 pKa = 11.84ARR76 pKa = 11.84ARR78 pKa = 11.84PRR80 pKa = 11.84RR81 pKa = 11.84HH82 pKa = 6.27LGGTPRR88 pKa = 11.84PSLAVFPRR96 pKa = 11.84PSLAVFPRR104 pKa = 11.84PNLAGPPRR112 pKa = 11.84GRR114 pKa = 11.84RR115 pKa = 11.84AGLPPRR121 pKa = 11.84ARR123 pKa = 11.84RR124 pKa = 11.84RR125 pKa = 11.84PRR127 pKa = 11.84TRR129 pKa = 11.84SDD131 pKa = 2.55RR132 pKa = 11.84RR133 pKa = 11.84RR134 pKa = 11.84AVLQAAVADD143 pKa = 4.29AGPIRR148 pKa = 11.84MNSSRR153 pKa = 11.84RR154 pKa = 11.84GARR157 pKa = 11.84AGTRR161 pKa = 11.84RR162 pKa = 11.84TPRR165 pKa = 11.84TARR168 pKa = 11.84RR169 pKa = 11.84PRR171 pKa = 11.84RR172 pKa = 11.84RR173 pKa = 11.84PSSPSPPSSRR183 pKa = 11.84GRR185 pKa = 11.84RR186 pKa = 11.84PGGWMPPIRR195 pKa = 11.84SRR197 pKa = 11.84GSGGVRR203 pKa = 11.84HH204 pKa = 6.83HH205 pKa = 6.64GRR207 pKa = 11.84PHH209 pKa = 6.5HH210 pKa = 5.71RR211 pKa = 11.84TAMAGSRR218 pKa = 11.84RR219 pKa = 11.84PVVPSTVPSTRR230 pKa = 11.84LPAGGPVRR238 pKa = 11.84SSPRR242 pKa = 11.84RR243 pKa = 11.84RR244 pKa = 11.84DD245 pKa = 2.88RR246 pKa = 11.84RR247 pKa = 11.84RR248 pKa = 11.84PLGRR252 pKa = 11.84VGTRR256 pKa = 11.84RR257 pKa = 11.84TVTNRR262 pKa = 11.84RR263 pKa = 11.84TRR265 pKa = 11.84IRR267 pKa = 11.84GRR269 pKa = 11.84ARR271 pKa = 11.84RR272 pKa = 11.84RR273 pKa = 3.23
MM1 pKa = 7.47SSRR4 pKa = 11.84RR5 pKa = 11.84SGTTTPPARR14 pKa = 11.84ALRR17 pKa = 11.84RR18 pKa = 11.84PRR20 pKa = 11.84TTSRR24 pKa = 11.84AGLRR28 pKa = 11.84RR29 pKa = 11.84VPTASPRR36 pKa = 11.84PVAGTLPAKK45 pKa = 10.39ARR47 pKa = 11.84TISVPAAQRR56 pKa = 11.84EE57 pKa = 4.45DD58 pKa = 2.99IRR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84APGRR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84ARR70 pKa = 11.84RR71 pKa = 11.84SRR73 pKa = 11.84RR74 pKa = 11.84ARR76 pKa = 11.84ARR78 pKa = 11.84PRR80 pKa = 11.84RR81 pKa = 11.84HH82 pKa = 6.27LGGTPRR88 pKa = 11.84PSLAVFPRR96 pKa = 11.84PSLAVFPRR104 pKa = 11.84PNLAGPPRR112 pKa = 11.84GRR114 pKa = 11.84RR115 pKa = 11.84AGLPPRR121 pKa = 11.84ARR123 pKa = 11.84RR124 pKa = 11.84RR125 pKa = 11.84PRR127 pKa = 11.84TRR129 pKa = 11.84SDD131 pKa = 2.55RR132 pKa = 11.84RR133 pKa = 11.84RR134 pKa = 11.84AVLQAAVADD143 pKa = 4.29AGPIRR148 pKa = 11.84MNSSRR153 pKa = 11.84RR154 pKa = 11.84GARR157 pKa = 11.84AGTRR161 pKa = 11.84RR162 pKa = 11.84TPRR165 pKa = 11.84TARR168 pKa = 11.84RR169 pKa = 11.84PRR171 pKa = 11.84RR172 pKa = 11.84RR173 pKa = 11.84PSSPSPPSSRR183 pKa = 11.84GRR185 pKa = 11.84RR186 pKa = 11.84PGGWMPPIRR195 pKa = 11.84SRR197 pKa = 11.84GSGGVRR203 pKa = 11.84HH204 pKa = 6.83HH205 pKa = 6.64GRR207 pKa = 11.84PHH209 pKa = 6.5HH210 pKa = 5.71RR211 pKa = 11.84TAMAGSRR218 pKa = 11.84RR219 pKa = 11.84PVVPSTVPSTRR230 pKa = 11.84LPAGGPVRR238 pKa = 11.84SSPRR242 pKa = 11.84RR243 pKa = 11.84RR244 pKa = 11.84DD245 pKa = 2.88RR246 pKa = 11.84RR247 pKa = 11.84RR248 pKa = 11.84PLGRR252 pKa = 11.84VGTRR256 pKa = 11.84RR257 pKa = 11.84TVTNRR262 pKa = 11.84RR263 pKa = 11.84TRR265 pKa = 11.84IRR267 pKa = 11.84GRR269 pKa = 11.84ARR271 pKa = 11.84RR272 pKa = 11.84RR273 pKa = 3.23
Molecular weight: 30.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1798018 |
29 |
8880 |
325.7 |
34.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.527 ± 0.053 | 0.761 ± 0.01 |
6.358 ± 0.027 | 5.869 ± 0.03 |
2.66 ± 0.019 | 9.396 ± 0.036 |
2.201 ± 0.015 | 3.305 ± 0.024 |
1.236 ± 0.023 | 10.412 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.669 ± 0.012 | 1.554 ± 0.017 |
6.186 ± 0.036 | 2.501 ± 0.018 |
8.867 ± 0.042 | 5.443 ± 0.023 |
5.975 ± 0.021 | 8.881 ± 0.032 |
1.475 ± 0.013 | 1.726 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
