
Sulfurospirillum deleyianum (strain ATCC 51133 / DSM 6946 / 5175)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Epsilonproteobacteria; Campylobacterales; Campylobacteraceae; Sulfurospirillum; Sulfurospirillum deleyianum
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
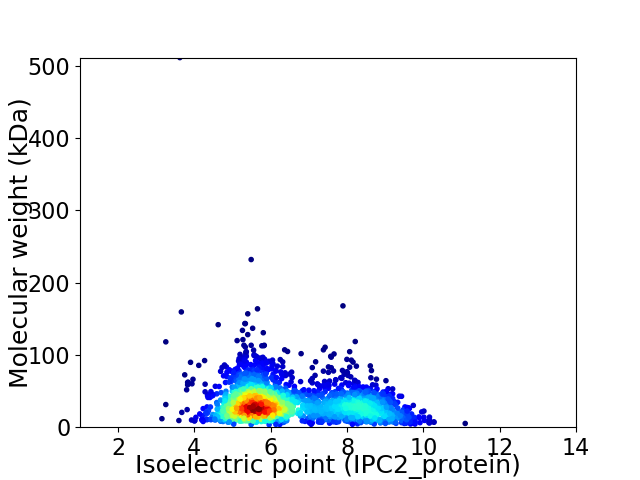
Virtual 2D-PAGE plot for 2243 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D1AYY0|D1AYY0_SULD5 Sulfatase OS=Sulfurospirillum deleyianum (strain ATCC 51133 / DSM 6946 / 5175) OX=525898 GN=Sdel_0080 PE=4 SV=1
MM1 pKa = 7.61NSSLYY6 pKa = 10.85NGVSGIKK13 pKa = 9.35TSQFYY18 pKa = 10.73IDD20 pKa = 4.3VIGNNISNVNTNGFKK35 pKa = 10.85GSTPEE40 pKa = 3.86VSSLFSSILVGTYY53 pKa = 9.85NSYY56 pKa = 11.54SNGVGMGAQSFNTALDD72 pKa = 3.55MSQGILEE79 pKa = 4.34NTDD82 pKa = 3.01NTFDD86 pKa = 4.16LALGSEE92 pKa = 4.56GWFGVQGADD101 pKa = 2.8GNTYY105 pKa = 7.16YY106 pKa = 10.54TRR108 pKa = 11.84AGSFSLDD115 pKa = 3.27AEE117 pKa = 4.7GNLVDD122 pKa = 5.15GDD124 pKa = 4.36GNYY127 pKa = 11.0LLATLGNNIATTTLDD142 pKa = 3.66ADD144 pKa = 3.95TLAKK148 pKa = 10.37FGQYY152 pKa = 10.3YY153 pKa = 10.92NNGTASSVTPYY164 pKa = 10.94AITEE168 pKa = 4.23LNDD171 pKa = 3.11VGLGTIGSQGIVNLPKK187 pKa = 9.51TLYY190 pKa = 8.69YY191 pKa = 10.09PPVASSEE198 pKa = 3.93ASYY201 pKa = 11.01SANLDD206 pKa = 3.35PTITTDD212 pKa = 5.41AVTLDD217 pKa = 4.74LNSADD222 pKa = 3.96YY223 pKa = 9.79PSSVLADD230 pKa = 3.74TTSKK234 pKa = 9.67TLTLNGTITNTITTQTPQKK253 pKa = 10.73GDD255 pKa = 3.34IVDD258 pKa = 3.77VTLKK262 pKa = 10.66DD263 pKa = 3.65ASGKK267 pKa = 8.11TLLLTATLDD276 pKa = 3.91EE277 pKa = 4.21NLEE280 pKa = 4.1WSINDD285 pKa = 3.68YY286 pKa = 11.07DD287 pKa = 4.06VSAYY291 pKa = 10.3DD292 pKa = 3.64LSSAFTLTSAKK303 pKa = 10.3LQTEE307 pKa = 4.33QEE309 pKa = 4.4VATQEE314 pKa = 4.0NFSTSIIGPDD324 pKa = 3.54GDD326 pKa = 5.17KK327 pKa = 11.03DD328 pKa = 3.61ILDD331 pKa = 3.47MTFTKK336 pKa = 10.37VIPQTSTEE344 pKa = 4.41TTWNAVVNIYY354 pKa = 10.72SFYY357 pKa = 9.86EE358 pKa = 4.15TYY360 pKa = 9.06DD361 pKa = 3.14TSKK364 pKa = 9.89TYY366 pKa = 9.19DD367 pKa = 3.13TTQYY371 pKa = 11.17YY372 pKa = 9.72IDD374 pKa = 3.4KK375 pKa = 8.97TANKK379 pKa = 9.42VYY381 pKa = 10.37EE382 pKa = 4.26IVDD385 pKa = 3.93TQTGALTFNGNGSLASNTIPTINNGGTTLTLNLGTVGGYY424 pKa = 10.78DD425 pKa = 3.68GMVSNANIDD434 pKa = 3.53KK435 pKa = 11.15ANASSSNGTLEE446 pKa = 4.47GFLTGYY452 pKa = 10.71DD453 pKa = 3.02VDD455 pKa = 4.15TNGNVIASFSNGKK468 pKa = 9.52SSSVAKK474 pKa = 9.78IAVYY478 pKa = 10.2HH479 pKa = 5.7FQNDD483 pKa = 3.25QGLMAEE489 pKa = 4.26SSNLFSEE496 pKa = 4.69SSNSGKK502 pKa = 10.57AFFYY506 pKa = 10.52ADD508 pKa = 3.4EE509 pKa = 4.67SGEE512 pKa = 4.2SFLGSKK518 pKa = 10.17VYY520 pKa = 9.49SHH522 pKa = 7.12KK523 pKa = 11.23LEE525 pKa = 4.27GSNVSLATALTEE537 pKa = 4.69LIIVQKK543 pKa = 11.07AFDD546 pKa = 4.07ASAKK550 pKa = 10.62SITTSDD556 pKa = 3.77EE557 pKa = 3.98LLKK560 pKa = 11.04NAINMKK566 pKa = 10.28AA567 pKa = 3.07
MM1 pKa = 7.61NSSLYY6 pKa = 10.85NGVSGIKK13 pKa = 9.35TSQFYY18 pKa = 10.73IDD20 pKa = 4.3VIGNNISNVNTNGFKK35 pKa = 10.85GSTPEE40 pKa = 3.86VSSLFSSILVGTYY53 pKa = 9.85NSYY56 pKa = 11.54SNGVGMGAQSFNTALDD72 pKa = 3.55MSQGILEE79 pKa = 4.34NTDD82 pKa = 3.01NTFDD86 pKa = 4.16LALGSEE92 pKa = 4.56GWFGVQGADD101 pKa = 2.8GNTYY105 pKa = 7.16YY106 pKa = 10.54TRR108 pKa = 11.84AGSFSLDD115 pKa = 3.27AEE117 pKa = 4.7GNLVDD122 pKa = 5.15GDD124 pKa = 4.36GNYY127 pKa = 11.0LLATLGNNIATTTLDD142 pKa = 3.66ADD144 pKa = 3.95TLAKK148 pKa = 10.37FGQYY152 pKa = 10.3YY153 pKa = 10.92NNGTASSVTPYY164 pKa = 10.94AITEE168 pKa = 4.23LNDD171 pKa = 3.11VGLGTIGSQGIVNLPKK187 pKa = 9.51TLYY190 pKa = 8.69YY191 pKa = 10.09PPVASSEE198 pKa = 3.93ASYY201 pKa = 11.01SANLDD206 pKa = 3.35PTITTDD212 pKa = 5.41AVTLDD217 pKa = 4.74LNSADD222 pKa = 3.96YY223 pKa = 9.79PSSVLADD230 pKa = 3.74TTSKK234 pKa = 9.67TLTLNGTITNTITTQTPQKK253 pKa = 10.73GDD255 pKa = 3.34IVDD258 pKa = 3.77VTLKK262 pKa = 10.66DD263 pKa = 3.65ASGKK267 pKa = 8.11TLLLTATLDD276 pKa = 3.91EE277 pKa = 4.21NLEE280 pKa = 4.1WSINDD285 pKa = 3.68YY286 pKa = 11.07DD287 pKa = 4.06VSAYY291 pKa = 10.3DD292 pKa = 3.64LSSAFTLTSAKK303 pKa = 10.3LQTEE307 pKa = 4.33QEE309 pKa = 4.4VATQEE314 pKa = 4.0NFSTSIIGPDD324 pKa = 3.54GDD326 pKa = 5.17KK327 pKa = 11.03DD328 pKa = 3.61ILDD331 pKa = 3.47MTFTKK336 pKa = 10.37VIPQTSTEE344 pKa = 4.41TTWNAVVNIYY354 pKa = 10.72SFYY357 pKa = 9.86EE358 pKa = 4.15TYY360 pKa = 9.06DD361 pKa = 3.14TSKK364 pKa = 9.89TYY366 pKa = 9.19DD367 pKa = 3.13TTQYY371 pKa = 11.17YY372 pKa = 9.72IDD374 pKa = 3.4KK375 pKa = 8.97TANKK379 pKa = 9.42VYY381 pKa = 10.37EE382 pKa = 4.26IVDD385 pKa = 3.93TQTGALTFNGNGSLASNTIPTINNGGTTLTLNLGTVGGYY424 pKa = 10.78DD425 pKa = 3.68GMVSNANIDD434 pKa = 3.53KK435 pKa = 11.15ANASSSNGTLEE446 pKa = 4.47GFLTGYY452 pKa = 10.71DD453 pKa = 3.02VDD455 pKa = 4.15TNGNVIASFSNGKK468 pKa = 9.52SSSVAKK474 pKa = 9.78IAVYY478 pKa = 10.2HH479 pKa = 5.7FQNDD483 pKa = 3.25QGLMAEE489 pKa = 4.26SSNLFSEE496 pKa = 4.69SSNSGKK502 pKa = 10.57AFFYY506 pKa = 10.52ADD508 pKa = 3.4EE509 pKa = 4.67SGEE512 pKa = 4.2SFLGSKK518 pKa = 10.17VYY520 pKa = 9.49SHH522 pKa = 7.12KK523 pKa = 11.23LEE525 pKa = 4.27GSNVSLATALTEE537 pKa = 4.69LIIVQKK543 pKa = 11.07AFDD546 pKa = 4.07ASAKK550 pKa = 10.62SITTSDD556 pKa = 3.77EE557 pKa = 3.98LLKK560 pKa = 11.04NAINMKK566 pKa = 10.28AA567 pKa = 3.07
Molecular weight: 60.0 kDa
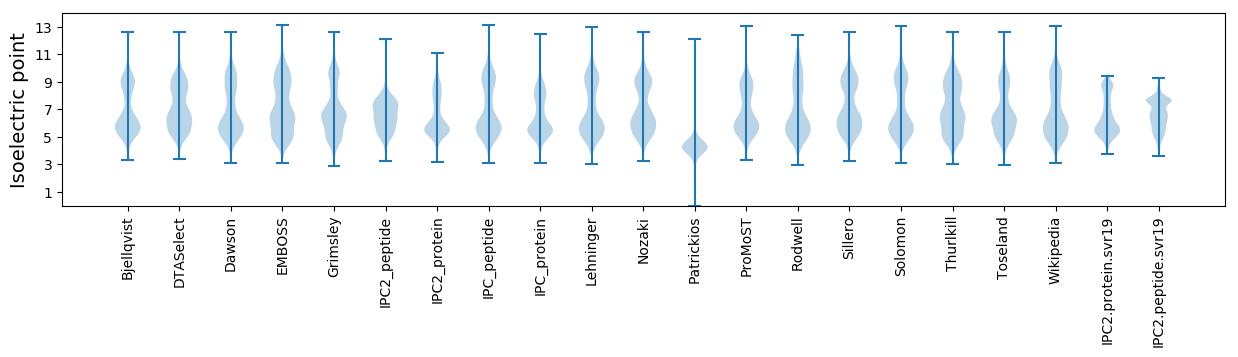
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D1B2Y5|D1B2Y5_SULD5 Uncharacterized protein OS=Sulfurospirillum deleyianum (strain ATCC 51133 / DSM 6946 / 5175) OX=525898 GN=Sdel_1437 PE=4 SV=1
MM1 pKa = 7.28KK2 pKa = 9.39RR3 pKa = 11.84TYY5 pKa = 10.1QPHH8 pKa = 4.92TTPKK12 pKa = 10.24KK13 pKa = 7.56RR14 pKa = 11.84THH16 pKa = 6.02GFRR19 pKa = 11.84LRR21 pKa = 11.84MKK23 pKa = 9.27TKK25 pKa = 10.31GGRR28 pKa = 11.84KK29 pKa = 9.04VIAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.08GRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84LAAA44 pKa = 4.25
MM1 pKa = 7.28KK2 pKa = 9.39RR3 pKa = 11.84TYY5 pKa = 10.1QPHH8 pKa = 4.92TTPKK12 pKa = 10.24KK13 pKa = 7.56RR14 pKa = 11.84THH16 pKa = 6.02GFRR19 pKa = 11.84LRR21 pKa = 11.84MKK23 pKa = 9.27TKK25 pKa = 10.31GGRR28 pKa = 11.84KK29 pKa = 9.04VIAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.08GRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84LAAA44 pKa = 4.25
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
701989 |
32 |
4798 |
313.0 |
35.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.492 ± 0.056 | 1.07 ± 0.023 |
4.945 ± 0.061 | 7.338 ± 0.05 |
5.047 ± 0.044 | 6.174 ± 0.064 |
2.379 ± 0.029 | 7.654 ± 0.045 |
7.603 ± 0.061 | 10.438 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.776 ± 0.029 | 4.431 ± 0.049 |
3.189 ± 0.036 | 3.172 ± 0.031 |
3.618 ± 0.035 | 6.556 ± 0.044 |
5.153 ± 0.051 | 6.457 ± 0.043 |
0.784 ± 0.019 | 3.723 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
