
Entomoplasma freundtii
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Tenericutes; Mollicutes; Entomoplasmatales; Entomoplasmataceae; Entomoplasma
Average proteome isoelectric point is 7.01
Get precalculated fractions of proteins
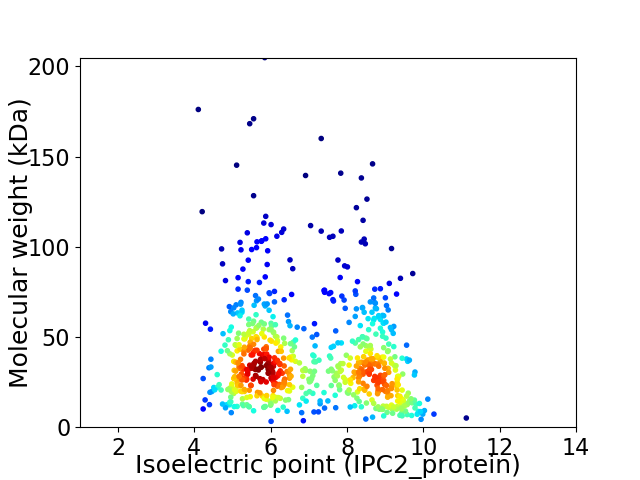
Virtual 2D-PAGE plot for 702 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2K8NRI7|A0A2K8NRI7_9MOLU Uncharacterized protein OS=Entomoplasma freundtii OX=74700 GN=EFREU_v1c04460 PE=4 SV=1
MM1 pKa = 7.48KK2 pKa = 10.49RR3 pKa = 11.84LLTLLTAIGFTTTTAVPVVACCKK26 pKa = 10.03TNTNNSGIKK35 pKa = 10.24DD36 pKa = 3.79LGTLQIRR43 pKa = 11.84QINLPIPKK51 pKa = 8.66ATSYY55 pKa = 10.4TSKK58 pKa = 10.29QANFVISMMTQNINDD73 pKa = 4.21QINEE77 pKa = 3.91DD78 pKa = 3.91TKK80 pKa = 11.21FKK82 pKa = 11.58NNIQNQDD89 pKa = 3.72MIDD92 pKa = 3.67YY93 pKa = 10.2TDD95 pKa = 5.21DD96 pKa = 3.42FTTNIADD103 pKa = 4.87FIKK106 pKa = 10.5PDD108 pKa = 3.61EE109 pKa = 4.25TVKK112 pKa = 10.37IDD114 pKa = 3.47GSGEE118 pKa = 3.69IQINISANQNSKK130 pKa = 10.27ILTGKK135 pKa = 8.38TNATIKK141 pKa = 10.45FNPNTQLASVINNTNLGEE159 pKa = 4.2FQTEE163 pKa = 4.01PTASDD168 pKa = 3.47IIGALSALNPQALIDD183 pKa = 3.94EE184 pKa = 5.11DD185 pKa = 4.18EE186 pKa = 4.83VEE188 pKa = 4.5VLASSINLNGATVKK202 pKa = 10.91AIDD205 pKa = 4.44SNNSLYY211 pKa = 10.61EE212 pKa = 3.72GQINVTYY219 pKa = 10.4KK220 pKa = 10.86VVGKK224 pKa = 9.75KK225 pKa = 8.71QLSEE229 pKa = 4.43VITNTSLGLVNDD241 pKa = 4.24ATNEE245 pKa = 4.25TILNAINQLNPEE257 pKa = 4.56ANLKK261 pKa = 7.21TTDD264 pKa = 3.25VTIVTNSANNGATITATEE282 pKa = 3.96TSNYY286 pKa = 9.06QGSIDD291 pKa = 3.43VTYY294 pKa = 9.75TIRR297 pKa = 11.84KK298 pKa = 7.42QLSTVVTNLDD308 pKa = 4.28LGNIDD313 pKa = 3.63NNANATILAAVNAKK327 pKa = 8.47NTDD330 pKa = 3.67ANLKK334 pKa = 9.39TSDD337 pKa = 3.24VQIAVNSDD345 pKa = 3.16NNGATLTATQSSLYY359 pKa = 9.24QGSVTVTFTVKK370 pKa = 9.35TALDD374 pKa = 3.69SVVKK378 pKa = 8.74VTALGNLDD386 pKa = 3.87NADD389 pKa = 4.58DD390 pKa = 4.05DD391 pKa = 4.87TILAAVNQANSGLKK405 pKa = 9.01LTTADD410 pKa = 3.48VSIKK414 pKa = 10.93VNDD417 pKa = 4.12DD418 pKa = 3.4KK419 pKa = 11.68KK420 pKa = 11.2SATLTANPDD429 pKa = 2.95GNYY432 pKa = 9.6QGSVTVTFTIKK443 pKa = 9.04ITLDD447 pKa = 3.43SVVKK451 pKa = 9.8VTDD454 pKa = 3.78LGKK457 pKa = 11.16LNDD460 pKa = 3.94TDD462 pKa = 4.62NNTILNAVNQVNSGLNLTNNDD483 pKa = 2.61VTIAINEE490 pKa = 4.23NKK492 pKa = 10.45DD493 pKa = 3.28GATLTATSSSLYY505 pKa = 9.68KK506 pKa = 10.84GSVTVTFTVKK516 pKa = 9.35TALDD520 pKa = 3.69SVVKK524 pKa = 8.74VTALGNLDD532 pKa = 3.87NADD535 pKa = 4.58DD536 pKa = 4.05DD537 pKa = 4.87TILAAVNQANSGLKK551 pKa = 9.01LTTADD556 pKa = 3.48VSIKK560 pKa = 10.91VNDD563 pKa = 4.11DD564 pKa = 3.42NKK566 pKa = 11.01SATLTANPDD575 pKa = 2.95GNYY578 pKa = 9.6QGSVTVTFTIKK589 pKa = 10.04IALDD593 pKa = 3.61SVVKK597 pKa = 9.83VTDD600 pKa = 3.78LGKK603 pKa = 11.12LNDD606 pKa = 4.21TDD608 pKa = 4.21NSTILNAVNQVNSGLNLTNNDD629 pKa = 2.61VTIAINEE636 pKa = 4.23NKK638 pKa = 10.45DD639 pKa = 3.28GATLTATPNGNYY651 pKa = 9.81KK652 pKa = 10.78GFVNVTFIVRR662 pKa = 11.84TQLSSVVTKK671 pKa = 10.49TNLGPIIKK679 pKa = 10.06ADD681 pKa = 3.11ATTILNAVNQVNSINLNASDD701 pKa = 5.32VDD703 pKa = 3.44IQNINNSSATLTSTSSSIYY722 pKa = 9.21QGSVTVTFTIKK733 pKa = 10.04IALDD737 pKa = 3.61SVVKK741 pKa = 9.79VTDD744 pKa = 4.41LGPIDD749 pKa = 4.3KK750 pKa = 10.6ADD752 pKa = 3.56SATILAEE759 pKa = 4.17VNSVNDD765 pKa = 4.3LNLTNNDD772 pKa = 3.05VTVVTNPDD780 pKa = 3.7DD781 pKa = 4.47KK782 pKa = 11.52SATLTANPDD791 pKa = 2.95GNYY794 pKa = 9.51QGSVEE799 pKa = 4.11VTFTIKK805 pKa = 9.92ATLEE809 pKa = 4.12LVVKK813 pKa = 9.53VTDD816 pKa = 4.66LGNLDD821 pKa = 3.94PVNDD825 pKa = 3.91EE826 pKa = 4.76TILDD830 pKa = 3.72AVNAKK835 pKa = 7.9NTNVSLKK842 pKa = 10.83ASDD845 pKa = 3.34VTIAIDD851 pKa = 3.78TDD853 pKa = 3.67NKK855 pKa = 10.81SATLTSTSSSIYY867 pKa = 9.21QGSVTVTFTIKK878 pKa = 10.04IALDD882 pKa = 3.61SVVKK886 pKa = 9.83VTDD889 pKa = 3.74LGKK892 pKa = 10.86LNEE895 pKa = 4.6ANNTTILAAVNAANKK910 pKa = 9.81GLNLTTNDD918 pKa = 2.91VTIIINVTQDD928 pKa = 3.28GANLTATEE936 pKa = 3.89TSIYY940 pKa = 9.24KK941 pKa = 10.53DD942 pKa = 3.4SVSVTFNIRR951 pKa = 11.84TQLSSVVTNLNLGNIDD967 pKa = 3.83NNSEE971 pKa = 3.95ATILNAVNAANGLKK985 pKa = 9.34LTTADD990 pKa = 3.48VSIKK994 pKa = 10.91VNDD997 pKa = 4.11DD998 pKa = 3.42NKK1000 pKa = 11.01SATLTANPDD1009 pKa = 2.91GNYY1012 pKa = 9.97QGLVTVTFTIKK1023 pKa = 10.04IALDD1027 pKa = 3.78SVVKK1031 pKa = 9.95ATDD1034 pKa = 3.42LGKK1037 pKa = 11.1LNDD1040 pKa = 4.21TDD1042 pKa = 3.85SSTILAAVNQVNSGLNLTTSDD1063 pKa = 3.13VTIKK1067 pKa = 10.9VNSNSSATITATEE1080 pKa = 3.89TSNYY1084 pKa = 8.67QGSVDD1089 pKa = 3.21VTFIVRR1095 pKa = 11.84TQLSSVVSKK1104 pKa = 10.17TNLGSINKK1112 pKa = 9.36ADD1114 pKa = 3.52STTILVAVNQVNGISLTTSDD1134 pKa = 4.32VDD1136 pKa = 3.4IQIIGNDD1143 pKa = 3.72SATLTATQSSLYY1155 pKa = 9.24QGSVTVTFTIKK1166 pKa = 10.04IALDD1170 pKa = 3.61SVVKK1174 pKa = 9.83VTDD1177 pKa = 3.73LGKK1180 pKa = 11.15LNDD1183 pKa = 4.29ADD1185 pKa = 4.09NNTILAAVNQVNSGLNLTTIDD1206 pKa = 3.19VTIKK1210 pKa = 10.75VNSNSSATLTAIEE1223 pKa = 4.09TSNYY1227 pKa = 8.7QGSVDD1232 pKa = 3.21VTFIVRR1238 pKa = 11.84TQLSSVVTNLDD1249 pKa = 3.78LGDD1252 pKa = 4.31IDD1254 pKa = 5.83NLTDD1258 pKa = 5.19DD1259 pKa = 5.2IILAAVNAKK1268 pKa = 9.99NPNANLKK1275 pKa = 10.68ASDD1278 pKa = 3.83VNIEE1282 pKa = 3.79ITGDD1286 pKa = 3.38NEE1288 pKa = 3.95ATLTATSTGVYY1299 pKa = 9.02QGSVTVTFTVRR1310 pKa = 11.84TQLSSVVTNRR1320 pKa = 11.84DD1321 pKa = 2.7LGAIDD1326 pKa = 5.36NLTDD1330 pKa = 5.05DD1331 pKa = 5.21IILAAVNAKK1340 pKa = 9.99NPNANLKK1347 pKa = 10.68ASDD1350 pKa = 3.85VNIEE1354 pKa = 4.05IIGDD1358 pKa = 3.71NEE1360 pKa = 3.99ATLTATQSSIYY1371 pKa = 9.24QGSVTVTFTIKK1382 pKa = 9.21PTLDD1386 pKa = 3.77SIVKK1390 pKa = 7.81VTNLGPIDD1398 pKa = 4.2KK1399 pKa = 10.61ADD1401 pKa = 3.58STNILAAVNVKK1412 pKa = 10.34NPSANLKK1419 pKa = 9.07DD1420 pKa = 3.5TDD1422 pKa = 3.52VTIQIGGSQDD1432 pKa = 3.29KK1433 pKa = 11.11AILTATKK1440 pKa = 9.51EE1441 pKa = 4.06SAYY1444 pKa = 10.11EE1445 pKa = 3.89GTVNVTFTIKK1455 pKa = 9.94ATLDD1459 pKa = 3.7SIVKK1463 pKa = 8.11VTALGILDD1471 pKa = 3.83NTDD1474 pKa = 4.3DD1475 pKa = 3.97EE1476 pKa = 5.18TILNAVNAVNKK1487 pKa = 10.04DD1488 pKa = 3.55FQLTSDD1494 pKa = 3.47EE1495 pKa = 4.16VTIVVNNDD1503 pKa = 2.48KK1504 pKa = 10.72TGATLTANPSSIYY1517 pKa = 9.92QGSVDD1522 pKa = 3.41VTFTIKK1528 pKa = 10.17QDD1530 pKa = 3.34LDD1532 pKa = 3.81TVVVDD1537 pKa = 4.99RR1538 pKa = 11.84NLGYY1542 pKa = 10.41INNNEE1547 pKa = 3.98LSTILEE1553 pKa = 4.25AVNLQNPNVNLKK1565 pKa = 8.57EE1566 pKa = 4.05TDD1568 pKa = 3.1VTIDD1572 pKa = 3.41VKK1574 pKa = 11.53GEE1576 pKa = 3.88NNATLTATQTSNYY1589 pKa = 8.4QGSVDD1594 pKa = 3.25VTFNIRR1600 pKa = 11.84EE1601 pKa = 4.07QLDD1604 pKa = 3.73SVITNFEE1611 pKa = 4.23LGEE1614 pKa = 4.19IDD1616 pKa = 3.55NTDD1619 pKa = 3.58PATIIAAVNAKK1630 pKa = 9.98NPNANLNVSDD1640 pKa = 3.5VTIVVNSNNTFATLTATKK1658 pKa = 10.45SSIYY1662 pKa = 9.47QGSVNVVFTIKK1673 pKa = 9.8TLL1675 pKa = 3.46
MM1 pKa = 7.48KK2 pKa = 10.49RR3 pKa = 11.84LLTLLTAIGFTTTTAVPVVACCKK26 pKa = 10.03TNTNNSGIKK35 pKa = 10.24DD36 pKa = 3.79LGTLQIRR43 pKa = 11.84QINLPIPKK51 pKa = 8.66ATSYY55 pKa = 10.4TSKK58 pKa = 10.29QANFVISMMTQNINDD73 pKa = 4.21QINEE77 pKa = 3.91DD78 pKa = 3.91TKK80 pKa = 11.21FKK82 pKa = 11.58NNIQNQDD89 pKa = 3.72MIDD92 pKa = 3.67YY93 pKa = 10.2TDD95 pKa = 5.21DD96 pKa = 3.42FTTNIADD103 pKa = 4.87FIKK106 pKa = 10.5PDD108 pKa = 3.61EE109 pKa = 4.25TVKK112 pKa = 10.37IDD114 pKa = 3.47GSGEE118 pKa = 3.69IQINISANQNSKK130 pKa = 10.27ILTGKK135 pKa = 8.38TNATIKK141 pKa = 10.45FNPNTQLASVINNTNLGEE159 pKa = 4.2FQTEE163 pKa = 4.01PTASDD168 pKa = 3.47IIGALSALNPQALIDD183 pKa = 3.94EE184 pKa = 5.11DD185 pKa = 4.18EE186 pKa = 4.83VEE188 pKa = 4.5VLASSINLNGATVKK202 pKa = 10.91AIDD205 pKa = 4.44SNNSLYY211 pKa = 10.61EE212 pKa = 3.72GQINVTYY219 pKa = 10.4KK220 pKa = 10.86VVGKK224 pKa = 9.75KK225 pKa = 8.71QLSEE229 pKa = 4.43VITNTSLGLVNDD241 pKa = 4.24ATNEE245 pKa = 4.25TILNAINQLNPEE257 pKa = 4.56ANLKK261 pKa = 7.21TTDD264 pKa = 3.25VTIVTNSANNGATITATEE282 pKa = 3.96TSNYY286 pKa = 9.06QGSIDD291 pKa = 3.43VTYY294 pKa = 9.75TIRR297 pKa = 11.84KK298 pKa = 7.42QLSTVVTNLDD308 pKa = 4.28LGNIDD313 pKa = 3.63NNANATILAAVNAKK327 pKa = 8.47NTDD330 pKa = 3.67ANLKK334 pKa = 9.39TSDD337 pKa = 3.24VQIAVNSDD345 pKa = 3.16NNGATLTATQSSLYY359 pKa = 9.24QGSVTVTFTVKK370 pKa = 9.35TALDD374 pKa = 3.69SVVKK378 pKa = 8.74VTALGNLDD386 pKa = 3.87NADD389 pKa = 4.58DD390 pKa = 4.05DD391 pKa = 4.87TILAAVNQANSGLKK405 pKa = 9.01LTTADD410 pKa = 3.48VSIKK414 pKa = 10.93VNDD417 pKa = 4.12DD418 pKa = 3.4KK419 pKa = 11.68KK420 pKa = 11.2SATLTANPDD429 pKa = 2.95GNYY432 pKa = 9.6QGSVTVTFTIKK443 pKa = 9.04ITLDD447 pKa = 3.43SVVKK451 pKa = 9.8VTDD454 pKa = 3.78LGKK457 pKa = 11.16LNDD460 pKa = 3.94TDD462 pKa = 4.62NNTILNAVNQVNSGLNLTNNDD483 pKa = 2.61VTIAINEE490 pKa = 4.23NKK492 pKa = 10.45DD493 pKa = 3.28GATLTATSSSLYY505 pKa = 9.68KK506 pKa = 10.84GSVTVTFTVKK516 pKa = 9.35TALDD520 pKa = 3.69SVVKK524 pKa = 8.74VTALGNLDD532 pKa = 3.87NADD535 pKa = 4.58DD536 pKa = 4.05DD537 pKa = 4.87TILAAVNQANSGLKK551 pKa = 9.01LTTADD556 pKa = 3.48VSIKK560 pKa = 10.91VNDD563 pKa = 4.11DD564 pKa = 3.42NKK566 pKa = 11.01SATLTANPDD575 pKa = 2.95GNYY578 pKa = 9.6QGSVTVTFTIKK589 pKa = 10.04IALDD593 pKa = 3.61SVVKK597 pKa = 9.83VTDD600 pKa = 3.78LGKK603 pKa = 11.12LNDD606 pKa = 4.21TDD608 pKa = 4.21NSTILNAVNQVNSGLNLTNNDD629 pKa = 2.61VTIAINEE636 pKa = 4.23NKK638 pKa = 10.45DD639 pKa = 3.28GATLTATPNGNYY651 pKa = 9.81KK652 pKa = 10.78GFVNVTFIVRR662 pKa = 11.84TQLSSVVTKK671 pKa = 10.49TNLGPIIKK679 pKa = 10.06ADD681 pKa = 3.11ATTILNAVNQVNSINLNASDD701 pKa = 5.32VDD703 pKa = 3.44IQNINNSSATLTSTSSSIYY722 pKa = 9.21QGSVTVTFTIKK733 pKa = 10.04IALDD737 pKa = 3.61SVVKK741 pKa = 9.79VTDD744 pKa = 4.41LGPIDD749 pKa = 4.3KK750 pKa = 10.6ADD752 pKa = 3.56SATILAEE759 pKa = 4.17VNSVNDD765 pKa = 4.3LNLTNNDD772 pKa = 3.05VTVVTNPDD780 pKa = 3.7DD781 pKa = 4.47KK782 pKa = 11.52SATLTANPDD791 pKa = 2.95GNYY794 pKa = 9.51QGSVEE799 pKa = 4.11VTFTIKK805 pKa = 9.92ATLEE809 pKa = 4.12LVVKK813 pKa = 9.53VTDD816 pKa = 4.66LGNLDD821 pKa = 3.94PVNDD825 pKa = 3.91EE826 pKa = 4.76TILDD830 pKa = 3.72AVNAKK835 pKa = 7.9NTNVSLKK842 pKa = 10.83ASDD845 pKa = 3.34VTIAIDD851 pKa = 3.78TDD853 pKa = 3.67NKK855 pKa = 10.81SATLTSTSSSIYY867 pKa = 9.21QGSVTVTFTIKK878 pKa = 10.04IALDD882 pKa = 3.61SVVKK886 pKa = 9.83VTDD889 pKa = 3.74LGKK892 pKa = 10.86LNEE895 pKa = 4.6ANNTTILAAVNAANKK910 pKa = 9.81GLNLTTNDD918 pKa = 2.91VTIIINVTQDD928 pKa = 3.28GANLTATEE936 pKa = 3.89TSIYY940 pKa = 9.24KK941 pKa = 10.53DD942 pKa = 3.4SVSVTFNIRR951 pKa = 11.84TQLSSVVTNLNLGNIDD967 pKa = 3.83NNSEE971 pKa = 3.95ATILNAVNAANGLKK985 pKa = 9.34LTTADD990 pKa = 3.48VSIKK994 pKa = 10.91VNDD997 pKa = 4.11DD998 pKa = 3.42NKK1000 pKa = 11.01SATLTANPDD1009 pKa = 2.91GNYY1012 pKa = 9.97QGLVTVTFTIKK1023 pKa = 10.04IALDD1027 pKa = 3.78SVVKK1031 pKa = 9.95ATDD1034 pKa = 3.42LGKK1037 pKa = 11.1LNDD1040 pKa = 4.21TDD1042 pKa = 3.85SSTILAAVNQVNSGLNLTTSDD1063 pKa = 3.13VTIKK1067 pKa = 10.9VNSNSSATITATEE1080 pKa = 3.89TSNYY1084 pKa = 8.67QGSVDD1089 pKa = 3.21VTFIVRR1095 pKa = 11.84TQLSSVVSKK1104 pKa = 10.17TNLGSINKK1112 pKa = 9.36ADD1114 pKa = 3.52STTILVAVNQVNGISLTTSDD1134 pKa = 4.32VDD1136 pKa = 3.4IQIIGNDD1143 pKa = 3.72SATLTATQSSLYY1155 pKa = 9.24QGSVTVTFTIKK1166 pKa = 10.04IALDD1170 pKa = 3.61SVVKK1174 pKa = 9.83VTDD1177 pKa = 3.73LGKK1180 pKa = 11.15LNDD1183 pKa = 4.29ADD1185 pKa = 4.09NNTILAAVNQVNSGLNLTTIDD1206 pKa = 3.19VTIKK1210 pKa = 10.75VNSNSSATLTAIEE1223 pKa = 4.09TSNYY1227 pKa = 8.7QGSVDD1232 pKa = 3.21VTFIVRR1238 pKa = 11.84TQLSSVVTNLDD1249 pKa = 3.78LGDD1252 pKa = 4.31IDD1254 pKa = 5.83NLTDD1258 pKa = 5.19DD1259 pKa = 5.2IILAAVNAKK1268 pKa = 9.99NPNANLKK1275 pKa = 10.68ASDD1278 pKa = 3.83VNIEE1282 pKa = 3.79ITGDD1286 pKa = 3.38NEE1288 pKa = 3.95ATLTATSTGVYY1299 pKa = 9.02QGSVTVTFTVRR1310 pKa = 11.84TQLSSVVTNRR1320 pKa = 11.84DD1321 pKa = 2.7LGAIDD1326 pKa = 5.36NLTDD1330 pKa = 5.05DD1331 pKa = 5.21IILAAVNAKK1340 pKa = 9.99NPNANLKK1347 pKa = 10.68ASDD1350 pKa = 3.85VNIEE1354 pKa = 4.05IIGDD1358 pKa = 3.71NEE1360 pKa = 3.99ATLTATQSSIYY1371 pKa = 9.24QGSVTVTFTIKK1382 pKa = 9.21PTLDD1386 pKa = 3.77SIVKK1390 pKa = 7.81VTNLGPIDD1398 pKa = 4.2KK1399 pKa = 10.61ADD1401 pKa = 3.58STNILAAVNVKK1412 pKa = 10.34NPSANLKK1419 pKa = 9.07DD1420 pKa = 3.5TDD1422 pKa = 3.52VTIQIGGSQDD1432 pKa = 3.29KK1433 pKa = 11.11AILTATKK1440 pKa = 9.51EE1441 pKa = 4.06SAYY1444 pKa = 10.11EE1445 pKa = 3.89GTVNVTFTIKK1455 pKa = 9.94ATLDD1459 pKa = 3.7SIVKK1463 pKa = 8.11VTALGILDD1471 pKa = 3.83NTDD1474 pKa = 4.3DD1475 pKa = 3.97EE1476 pKa = 5.18TILNAVNAVNKK1487 pKa = 10.04DD1488 pKa = 3.55FQLTSDD1494 pKa = 3.47EE1495 pKa = 4.16VTIVVNNDD1503 pKa = 2.48KK1504 pKa = 10.72TGATLTANPSSIYY1517 pKa = 9.92QGSVDD1522 pKa = 3.41VTFTIKK1528 pKa = 10.17QDD1530 pKa = 3.34LDD1532 pKa = 3.81TVVVDD1537 pKa = 4.99RR1538 pKa = 11.84NLGYY1542 pKa = 10.41INNNEE1547 pKa = 3.98LSTILEE1553 pKa = 4.25AVNLQNPNVNLKK1565 pKa = 8.57EE1566 pKa = 4.05TDD1568 pKa = 3.1VTIDD1572 pKa = 3.41VKK1574 pKa = 11.53GEE1576 pKa = 3.88NNATLTATQTSNYY1589 pKa = 8.4QGSVDD1594 pKa = 3.25VTFNIRR1600 pKa = 11.84EE1601 pKa = 4.07QLDD1604 pKa = 3.73SVITNFEE1611 pKa = 4.23LGEE1614 pKa = 4.19IDD1616 pKa = 3.55NTDD1619 pKa = 3.58PATIIAAVNAKK1630 pKa = 9.98NPNANLNVSDD1640 pKa = 3.5VTIVVNSNNTFATLTATKK1658 pKa = 10.45SSIYY1662 pKa = 9.47QGSVNVVFTIKK1673 pKa = 9.8TLL1675 pKa = 3.46
Molecular weight: 176.1 kDa
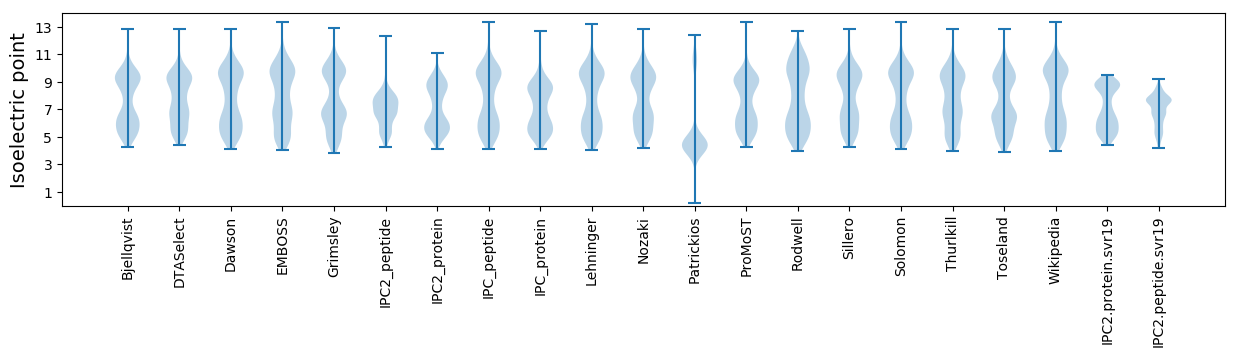
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2K8NSF9|A0A2K8NSF9_9MOLU Single-stranded DNA-binding protein OS=Entomoplasma freundtii OX=74700 GN=ssb PE=3 SV=1
MM1 pKa = 7.32KK2 pKa = 9.47RR3 pKa = 11.84TWQPSKK9 pKa = 10.03IKK11 pKa = 10.14HH12 pKa = 5.53ARR14 pKa = 11.84THH16 pKa = 5.78GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.41NGRR28 pKa = 11.84KK29 pKa = 9.2VIKK32 pKa = 10.14ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.34GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
MM1 pKa = 7.32KK2 pKa = 9.47RR3 pKa = 11.84TWQPSKK9 pKa = 10.03IKK11 pKa = 10.14HH12 pKa = 5.53ARR14 pKa = 11.84THH16 pKa = 5.78GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.41NGRR28 pKa = 11.84KK29 pKa = 9.2VIKK32 pKa = 10.14ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.34GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
251788 |
30 |
1816 |
358.7 |
40.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.467 ± 0.077 | 0.594 ± 0.026 |
5.246 ± 0.076 | 6.155 ± 0.097 |
4.967 ± 0.085 | 5.915 ± 0.081 |
1.825 ± 0.05 | 8.077 ± 0.084 |
8.264 ± 0.075 | 10.435 ± 0.093 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.225 ± 0.041 | 6.291 ± 0.091 |
3.579 ± 0.044 | 4.112 ± 0.056 |
3.372 ± 0.054 | 5.668 ± 0.07 |
5.867 ± 0.083 | 6.112 ± 0.071 |
1.282 ± 0.037 | 3.547 ± 0.05 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
