
Stella humosa
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Stellaceae; Stella
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
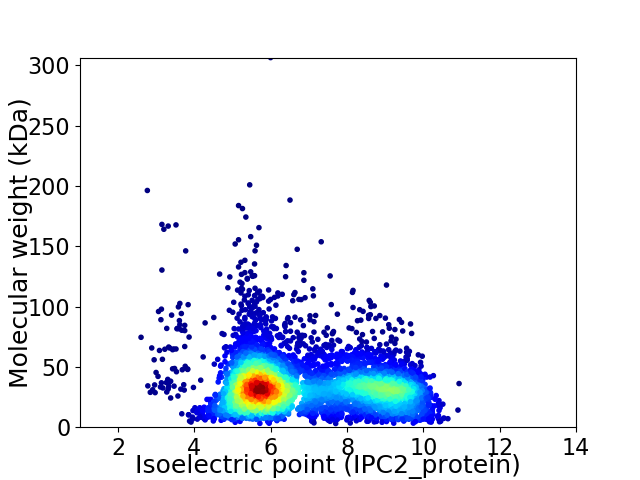
Virtual 2D-PAGE plot for 5422 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3N1M9X5|A0A3N1M9X5_9PROT Uncharacterized protein OS=Stella humosa OX=94 GN=EDC65_2270 PE=4 SV=1
MM1 pKa = 7.28YY2 pKa = 8.3ATGNYY7 pKa = 9.48AIAVDD12 pKa = 4.96AYY14 pKa = 10.95NLYY17 pKa = 10.63APPPPPPPPPPPPPPSPHH35 pKa = 4.42QTINGQAGPDD45 pKa = 4.17SIDD48 pKa = 3.72GDD50 pKa = 3.71WGNDD54 pKa = 3.56TIRR57 pKa = 11.84GYY59 pKa = 10.76SGNDD63 pKa = 3.45TIRR66 pKa = 11.84GLDD69 pKa = 3.93GNDD72 pKa = 3.54KK73 pKa = 11.07LYY75 pKa = 11.24GDD77 pKa = 5.67DD78 pKa = 5.76GDD80 pKa = 5.81DD81 pKa = 4.49DD82 pKa = 5.21VNGNLGNDD90 pKa = 3.86LVHH93 pKa = 6.97GGPGQDD99 pKa = 3.74FVRR102 pKa = 11.84GGQGNDD108 pKa = 3.07QVFGQDD114 pKa = 2.91GDD116 pKa = 3.84DD117 pKa = 3.08WHH119 pKa = 7.87VNGNIGNDD127 pKa = 3.26SVYY130 pKa = 11.01GGTGNDD136 pKa = 3.77GVFGGPGQDD145 pKa = 2.99TLYY148 pKa = 11.37GEE150 pKa = 5.01AGNDD154 pKa = 3.59TLSGDD159 pKa = 4.21LEE161 pKa = 4.57NDD163 pKa = 3.25ALYY166 pKa = 10.63GGPGADD172 pKa = 3.21LFVINSNGGWDD183 pKa = 3.62VIGDD187 pKa = 4.01FNRR190 pKa = 11.84SQGDD194 pKa = 3.68KK195 pKa = 10.66IVIQADD201 pKa = 3.28INGTGVFTGQQVLAMVTTGGGGAALIDD228 pKa = 3.83FGGGHH233 pKa = 7.1KK234 pKa = 10.52LGILGVSPGQLRR246 pKa = 11.84ADD248 pKa = 4.49DD249 pKa = 4.06FLIMM253 pKa = 5.02
MM1 pKa = 7.28YY2 pKa = 8.3ATGNYY7 pKa = 9.48AIAVDD12 pKa = 4.96AYY14 pKa = 10.95NLYY17 pKa = 10.63APPPPPPPPPPPPPPSPHH35 pKa = 4.42QTINGQAGPDD45 pKa = 4.17SIDD48 pKa = 3.72GDD50 pKa = 3.71WGNDD54 pKa = 3.56TIRR57 pKa = 11.84GYY59 pKa = 10.76SGNDD63 pKa = 3.45TIRR66 pKa = 11.84GLDD69 pKa = 3.93GNDD72 pKa = 3.54KK73 pKa = 11.07LYY75 pKa = 11.24GDD77 pKa = 5.67DD78 pKa = 5.76GDD80 pKa = 5.81DD81 pKa = 4.49DD82 pKa = 5.21VNGNLGNDD90 pKa = 3.86LVHH93 pKa = 6.97GGPGQDD99 pKa = 3.74FVRR102 pKa = 11.84GGQGNDD108 pKa = 3.07QVFGQDD114 pKa = 2.91GDD116 pKa = 3.84DD117 pKa = 3.08WHH119 pKa = 7.87VNGNIGNDD127 pKa = 3.26SVYY130 pKa = 11.01GGTGNDD136 pKa = 3.77GVFGGPGQDD145 pKa = 2.99TLYY148 pKa = 11.37GEE150 pKa = 5.01AGNDD154 pKa = 3.59TLSGDD159 pKa = 4.21LEE161 pKa = 4.57NDD163 pKa = 3.25ALYY166 pKa = 10.63GGPGADD172 pKa = 3.21LFVINSNGGWDD183 pKa = 3.62VIGDD187 pKa = 4.01FNRR190 pKa = 11.84SQGDD194 pKa = 3.68KK195 pKa = 10.66IVIQADD201 pKa = 3.28INGTGVFTGQQVLAMVTTGGGGAALIDD228 pKa = 3.83FGGGHH233 pKa = 7.1KK234 pKa = 10.52LGILGVSPGQLRR246 pKa = 11.84ADD248 pKa = 4.49DD249 pKa = 4.06FLIMM253 pKa = 5.02
Molecular weight: 25.86 kDa
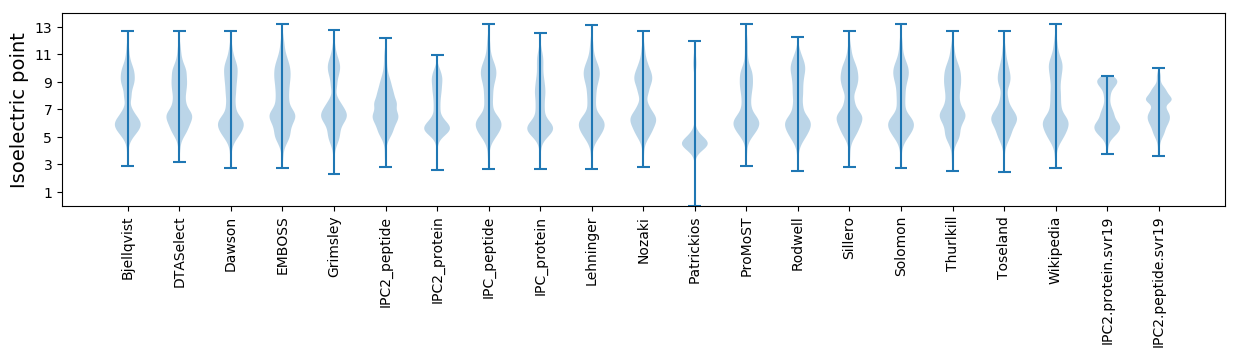
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3N1KWV8|A0A3N1KWV8_9PROT Uncharacterized protein OS=Stella humosa OX=94 GN=EDC65_4792 PE=4 SV=1
MM1 pKa = 7.78RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84PPRR7 pKa = 11.84WPRR10 pKa = 11.84PSPTTRR16 pKa = 11.84IRR18 pKa = 11.84IAVAASAASAARR30 pKa = 11.84KK31 pKa = 9.37ASSRR35 pKa = 11.84AILRR39 pKa = 11.84RR40 pKa = 11.84LRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84ARR46 pKa = 11.84PSPSEE51 pKa = 3.63ARR53 pKa = 11.84TPSVPSARR61 pKa = 11.84AAQRR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84SASRR71 pKa = 11.84SRR73 pKa = 11.84SANRR77 pKa = 11.84PSPSPPRR84 pKa = 11.84ARR86 pKa = 11.84PSRR89 pKa = 11.84RR90 pKa = 11.84QPSAGRR96 pKa = 11.84NRR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84PSRR103 pKa = 11.84RR104 pKa = 11.84QHH106 pKa = 6.29RR107 pKa = 11.84SASRR111 pKa = 11.84SRR113 pKa = 11.84LPPGRR118 pKa = 11.84RR119 pKa = 11.84HH120 pKa = 6.01LPLRR124 pKa = 11.84PRR126 pKa = 11.84RR127 pKa = 11.84RR128 pKa = 11.84SGRR131 pKa = 11.84ASVKK135 pKa = 10.4DD136 pKa = 3.68GQSVKK141 pKa = 10.36DD142 pKa = 3.88GQSARR147 pKa = 11.84PGQSATRR154 pKa = 11.84PHH156 pKa = 6.72RR157 pKa = 11.84RR158 pKa = 11.84PPASARR164 pKa = 11.84HH165 pKa = 5.26SRR167 pKa = 11.84ATPAAGRR174 pKa = 11.84KK175 pKa = 8.3PNPNSSARR183 pKa = 11.84RR184 pKa = 11.84PDD186 pKa = 3.25RR187 pKa = 11.84AMLADD192 pKa = 3.93RR193 pKa = 11.84CRR195 pKa = 11.84SRR197 pKa = 11.84RR198 pKa = 11.84RR199 pKa = 11.84PPNARR204 pKa = 11.84RR205 pKa = 11.84NDD207 pKa = 3.54RR208 pKa = 11.84PRR210 pKa = 11.84RR211 pKa = 11.84RR212 pKa = 11.84HH213 pKa = 4.89PRR215 pKa = 11.84PRR217 pKa = 11.84CSVLANAMPVRR228 pKa = 11.84ASRR231 pKa = 11.84HH232 pKa = 4.43PRR234 pKa = 11.84ATGTRR239 pKa = 11.84ATAIGSRR246 pKa = 11.84HH247 pKa = 6.2PPTDD251 pKa = 2.78RR252 pKa = 11.84RR253 pKa = 11.84RR254 pKa = 11.84AGRR257 pKa = 11.84PSAARR262 pKa = 11.84ASRR265 pKa = 11.84TGEE268 pKa = 3.76ASPTARR274 pKa = 11.84ACRR277 pKa = 11.84PTPPSIRR284 pKa = 11.84PGRR287 pKa = 11.84ACHH290 pKa = 6.15RR291 pKa = 11.84PWWHH295 pKa = 6.95LARR298 pKa = 11.84PRR300 pKa = 11.84QTSCVHH306 pKa = 6.4AAWTMSAAAAPRR318 pKa = 11.84ARR320 pKa = 11.84KK321 pKa = 8.66AAAPP325 pKa = 3.78
MM1 pKa = 7.78RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84PPRR7 pKa = 11.84WPRR10 pKa = 11.84PSPTTRR16 pKa = 11.84IRR18 pKa = 11.84IAVAASAASAARR30 pKa = 11.84KK31 pKa = 9.37ASSRR35 pKa = 11.84AILRR39 pKa = 11.84RR40 pKa = 11.84LRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84ARR46 pKa = 11.84PSPSEE51 pKa = 3.63ARR53 pKa = 11.84TPSVPSARR61 pKa = 11.84AAQRR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84SASRR71 pKa = 11.84SRR73 pKa = 11.84SANRR77 pKa = 11.84PSPSPPRR84 pKa = 11.84ARR86 pKa = 11.84PSRR89 pKa = 11.84RR90 pKa = 11.84QPSAGRR96 pKa = 11.84NRR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84PSRR103 pKa = 11.84RR104 pKa = 11.84QHH106 pKa = 6.29RR107 pKa = 11.84SASRR111 pKa = 11.84SRR113 pKa = 11.84LPPGRR118 pKa = 11.84RR119 pKa = 11.84HH120 pKa = 6.01LPLRR124 pKa = 11.84PRR126 pKa = 11.84RR127 pKa = 11.84RR128 pKa = 11.84SGRR131 pKa = 11.84ASVKK135 pKa = 10.4DD136 pKa = 3.68GQSVKK141 pKa = 10.36DD142 pKa = 3.88GQSARR147 pKa = 11.84PGQSATRR154 pKa = 11.84PHH156 pKa = 6.72RR157 pKa = 11.84RR158 pKa = 11.84PPASARR164 pKa = 11.84HH165 pKa = 5.26SRR167 pKa = 11.84ATPAAGRR174 pKa = 11.84KK175 pKa = 8.3PNPNSSARR183 pKa = 11.84RR184 pKa = 11.84PDD186 pKa = 3.25RR187 pKa = 11.84AMLADD192 pKa = 3.93RR193 pKa = 11.84CRR195 pKa = 11.84SRR197 pKa = 11.84RR198 pKa = 11.84RR199 pKa = 11.84PPNARR204 pKa = 11.84RR205 pKa = 11.84NDD207 pKa = 3.54RR208 pKa = 11.84PRR210 pKa = 11.84RR211 pKa = 11.84RR212 pKa = 11.84HH213 pKa = 4.89PRR215 pKa = 11.84PRR217 pKa = 11.84CSVLANAMPVRR228 pKa = 11.84ASRR231 pKa = 11.84HH232 pKa = 4.43PRR234 pKa = 11.84ATGTRR239 pKa = 11.84ATAIGSRR246 pKa = 11.84HH247 pKa = 6.2PPTDD251 pKa = 2.78RR252 pKa = 11.84RR253 pKa = 11.84RR254 pKa = 11.84AGRR257 pKa = 11.84PSAARR262 pKa = 11.84ASRR265 pKa = 11.84TGEE268 pKa = 3.76ASPTARR274 pKa = 11.84ACRR277 pKa = 11.84PTPPSIRR284 pKa = 11.84PGRR287 pKa = 11.84ACHH290 pKa = 6.15RR291 pKa = 11.84PWWHH295 pKa = 6.95LARR298 pKa = 11.84PRR300 pKa = 11.84QTSCVHH306 pKa = 6.4AAWTMSAAAAPRR318 pKa = 11.84ARR320 pKa = 11.84KK321 pKa = 8.66AAAPP325 pKa = 3.78
Molecular weight: 36.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1801500 |
29 |
2835 |
332.3 |
35.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.034 ± 0.051 | 0.799 ± 0.01 |
5.831 ± 0.041 | 4.991 ± 0.03 |
3.533 ± 0.021 | 9.489 ± 0.05 |
2.024 ± 0.017 | 4.718 ± 0.021 |
2.194 ± 0.027 | 10.341 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.466 ± 0.015 | 2.084 ± 0.018 |
5.783 ± 0.03 | 2.851 ± 0.018 |
8.005 ± 0.04 | 4.447 ± 0.023 |
5.048 ± 0.021 | 7.849 ± 0.028 |
1.45 ± 0.014 | 2.063 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
