
Treponema primitia (strain ATCC BAA-887 / DSM 12427 / ZAS-2)
Taxonomy: cellular organisms; Bacteria; Spirochaetes; Spirochaetia; Spirochaetales; Treponemataceae; Treponema; Treponema primitia
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
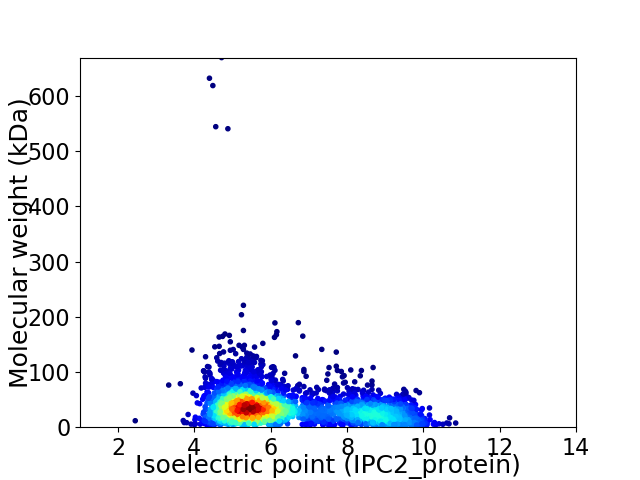
Virtual 2D-PAGE plot for 3512 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
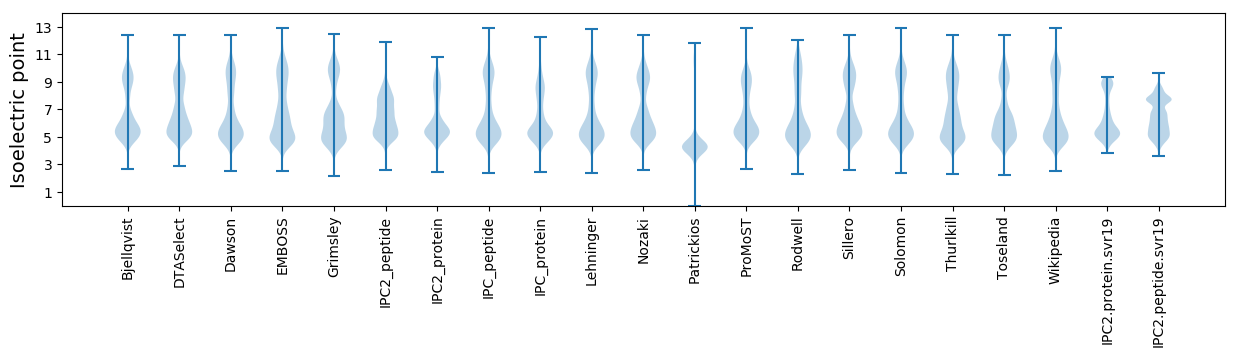
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F5YRI1|F5YRI1_TREPZ Uncharacterized protein OS=Treponema primitia (strain ATCC BAA-887 / DSM 12427 / ZAS-2) OX=545694 GN=TREPR_1248 PE=4 SV=1
MM1 pKa = 7.57KK2 pKa = 10.72LFGNKK7 pKa = 7.39QRR9 pKa = 11.84ALSPITCAAGIVLLAGLLALTGCANPLNSPADD41 pKa = 3.6NTVGQGLGLVTITIGEE57 pKa = 4.12TGARR61 pKa = 11.84NLYY64 pKa = 9.41PNPTGFTNYY73 pKa = 9.28EE74 pKa = 3.86LSFAPISGPAFYY86 pKa = 10.41GAIPASVAKK95 pKa = 9.93SDD97 pKa = 3.87SSFPPISLIPGTWTITAKK115 pKa = 10.45AYY117 pKa = 10.67NGSTQVAEE125 pKa = 4.24TAVEE129 pKa = 4.28VTVVAGEE136 pKa = 4.33LVTKK140 pKa = 10.63NIILKK145 pKa = 9.67PITGAGAGTFDD156 pKa = 4.73YY157 pKa = 10.85EE158 pKa = 4.29VTNADD163 pKa = 5.13DD164 pKa = 3.94VDD166 pKa = 4.4FDD168 pKa = 5.12DD169 pKa = 6.33GSEE172 pKa = 4.11LTLSDD177 pKa = 4.26LSGNPIYY184 pKa = 10.19TYY186 pKa = 10.92NLSSVQSGTDD196 pKa = 3.34SVPAGIYY203 pKa = 10.11DD204 pKa = 3.74LSIVIKK210 pKa = 10.23KK211 pKa = 10.56DD212 pKa = 3.13GGITGTTEE220 pKa = 3.75VVFIYY225 pKa = 10.39PGLTTVAHH233 pKa = 6.82FIFGNGEE240 pKa = 4.01IYY242 pKa = 10.7SGTTSNPAFVATSLSKK258 pKa = 10.08TEE260 pKa = 4.04VVQPSITFEE269 pKa = 4.28LKK271 pKa = 10.58NDD273 pKa = 3.86TEE275 pKa = 4.5LTGATWKK282 pKa = 10.93VYY284 pKa = 10.18DD285 pKa = 5.01AEE287 pKa = 5.99DD288 pKa = 4.64DD289 pKa = 4.18GDD291 pKa = 5.53DD292 pKa = 3.5ITDD295 pKa = 3.67TVVPTIEE302 pKa = 4.39SSNKK306 pKa = 8.36LTLTAVSNNITAGDD320 pKa = 3.77YY321 pKa = 9.27WVTAQAPGKK330 pKa = 9.78FEE332 pKa = 3.95SATRR336 pKa = 11.84VKK338 pKa = 10.22LTVEE342 pKa = 4.06PNPLQNEE349 pKa = 4.44TFTVVDD355 pKa = 4.97GIGDD359 pKa = 3.8PGLNNIYY366 pKa = 10.71LEE368 pKa = 4.39GGFVKK373 pKa = 10.43IGDD376 pKa = 4.2DD377 pKa = 3.45DD378 pKa = 5.05GGFIDD383 pKa = 5.5HH384 pKa = 7.07NPDD387 pKa = 2.86TDD389 pKa = 3.72PAYY392 pKa = 10.03KK393 pKa = 10.09PLAVFLGLTGVEE405 pKa = 4.31PDD407 pKa = 3.58DD408 pKa = 4.4QIVFDD413 pKa = 4.86EE414 pKa = 4.3YY415 pKa = 11.82GKK417 pKa = 10.06ILKK420 pKa = 8.04LTKK423 pKa = 10.17VDD425 pKa = 2.99ITYY428 pKa = 10.4DD429 pKa = 2.9ITFAVDD435 pKa = 4.29DD436 pKa = 4.85KK437 pKa = 10.82YY438 pKa = 11.6LPAIADD444 pKa = 3.77VLTLTGNASKK454 pKa = 10.73SFEE457 pKa = 4.24VTGDD461 pKa = 3.33ALTGKK466 pKa = 7.74TVILDD471 pKa = 3.79STSSPTYY478 pKa = 8.98NFYY481 pKa = 10.97GVSVLPYY488 pKa = 8.72DD489 pKa = 4.1TNDD492 pKa = 3.19VTISAEE498 pKa = 4.92DD499 pKa = 4.32DD500 pKa = 3.59DD501 pKa = 4.69TDD503 pKa = 4.01GKK505 pKa = 11.23VVTLLDD511 pKa = 4.19GGSADD516 pKa = 3.47ATVSVLDD523 pKa = 3.76EE524 pKa = 4.57AVDD527 pKa = 3.73KK528 pKa = 11.32LVLGTNAPSAISNEE542 pKa = 3.77KK543 pKa = 9.97SGLVIDD549 pKa = 4.28VGTNAGITITATQPTTVVTDD569 pKa = 3.75KK570 pKa = 11.06LATGTVTVDD579 pKa = 3.99GYY581 pKa = 9.63STAAITIDD589 pKa = 3.62GVSITPVGYY598 pKa = 9.24TDD600 pKa = 6.04AIVTKK605 pKa = 9.44TDD607 pKa = 3.45YY608 pKa = 9.88YY609 pKa = 10.89TLEE612 pKa = 5.11LDD614 pKa = 5.02AGDD617 pKa = 3.79TTATVAIEE625 pKa = 3.95YY626 pKa = 10.43DD627 pKa = 3.63SAITEE632 pKa = 4.19LTLVDD637 pKa = 4.04TNLEE641 pKa = 4.46AISNANTATVINVGSISGVEE661 pKa = 3.46ITAAVDD667 pKa = 3.43TTVVTTEE674 pKa = 4.38LSDD677 pKa = 3.72SNTVVLIGDD686 pKa = 3.6KK687 pKa = 10.76TFTLLVLDD695 pKa = 4.67DD696 pKa = 5.18GGYY699 pKa = 10.86EE700 pKa = 4.49DD701 pKa = 6.13DD702 pKa = 4.3PTPGTVTGSTNGYY715 pKa = 9.79VYY717 pKa = 10.38TYY719 pKa = 11.15LNDD722 pKa = 3.58NGNEE726 pKa = 4.0NTEE729 pKa = 4.13FAPIDD734 pKa = 4.19DD735 pKa = 5.37DD736 pKa = 5.11SFSLTSDD743 pKa = 3.02SGTVTVTVSDD753 pKa = 3.53ITPP756 pKa = 3.46
MM1 pKa = 7.57KK2 pKa = 10.72LFGNKK7 pKa = 7.39QRR9 pKa = 11.84ALSPITCAAGIVLLAGLLALTGCANPLNSPADD41 pKa = 3.6NTVGQGLGLVTITIGEE57 pKa = 4.12TGARR61 pKa = 11.84NLYY64 pKa = 9.41PNPTGFTNYY73 pKa = 9.28EE74 pKa = 3.86LSFAPISGPAFYY86 pKa = 10.41GAIPASVAKK95 pKa = 9.93SDD97 pKa = 3.87SSFPPISLIPGTWTITAKK115 pKa = 10.45AYY117 pKa = 10.67NGSTQVAEE125 pKa = 4.24TAVEE129 pKa = 4.28VTVVAGEE136 pKa = 4.33LVTKK140 pKa = 10.63NIILKK145 pKa = 9.67PITGAGAGTFDD156 pKa = 4.73YY157 pKa = 10.85EE158 pKa = 4.29VTNADD163 pKa = 5.13DD164 pKa = 3.94VDD166 pKa = 4.4FDD168 pKa = 5.12DD169 pKa = 6.33GSEE172 pKa = 4.11LTLSDD177 pKa = 4.26LSGNPIYY184 pKa = 10.19TYY186 pKa = 10.92NLSSVQSGTDD196 pKa = 3.34SVPAGIYY203 pKa = 10.11DD204 pKa = 3.74LSIVIKK210 pKa = 10.23KK211 pKa = 10.56DD212 pKa = 3.13GGITGTTEE220 pKa = 3.75VVFIYY225 pKa = 10.39PGLTTVAHH233 pKa = 6.82FIFGNGEE240 pKa = 4.01IYY242 pKa = 10.7SGTTSNPAFVATSLSKK258 pKa = 10.08TEE260 pKa = 4.04VVQPSITFEE269 pKa = 4.28LKK271 pKa = 10.58NDD273 pKa = 3.86TEE275 pKa = 4.5LTGATWKK282 pKa = 10.93VYY284 pKa = 10.18DD285 pKa = 5.01AEE287 pKa = 5.99DD288 pKa = 4.64DD289 pKa = 4.18GDD291 pKa = 5.53DD292 pKa = 3.5ITDD295 pKa = 3.67TVVPTIEE302 pKa = 4.39SSNKK306 pKa = 8.36LTLTAVSNNITAGDD320 pKa = 3.77YY321 pKa = 9.27WVTAQAPGKK330 pKa = 9.78FEE332 pKa = 3.95SATRR336 pKa = 11.84VKK338 pKa = 10.22LTVEE342 pKa = 4.06PNPLQNEE349 pKa = 4.44TFTVVDD355 pKa = 4.97GIGDD359 pKa = 3.8PGLNNIYY366 pKa = 10.71LEE368 pKa = 4.39GGFVKK373 pKa = 10.43IGDD376 pKa = 4.2DD377 pKa = 3.45DD378 pKa = 5.05GGFIDD383 pKa = 5.5HH384 pKa = 7.07NPDD387 pKa = 2.86TDD389 pKa = 3.72PAYY392 pKa = 10.03KK393 pKa = 10.09PLAVFLGLTGVEE405 pKa = 4.31PDD407 pKa = 3.58DD408 pKa = 4.4QIVFDD413 pKa = 4.86EE414 pKa = 4.3YY415 pKa = 11.82GKK417 pKa = 10.06ILKK420 pKa = 8.04LTKK423 pKa = 10.17VDD425 pKa = 2.99ITYY428 pKa = 10.4DD429 pKa = 2.9ITFAVDD435 pKa = 4.29DD436 pKa = 4.85KK437 pKa = 10.82YY438 pKa = 11.6LPAIADD444 pKa = 3.77VLTLTGNASKK454 pKa = 10.73SFEE457 pKa = 4.24VTGDD461 pKa = 3.33ALTGKK466 pKa = 7.74TVILDD471 pKa = 3.79STSSPTYY478 pKa = 8.98NFYY481 pKa = 10.97GVSVLPYY488 pKa = 8.72DD489 pKa = 4.1TNDD492 pKa = 3.19VTISAEE498 pKa = 4.92DD499 pKa = 4.32DD500 pKa = 3.59DD501 pKa = 4.69TDD503 pKa = 4.01GKK505 pKa = 11.23VVTLLDD511 pKa = 4.19GGSADD516 pKa = 3.47ATVSVLDD523 pKa = 3.76EE524 pKa = 4.57AVDD527 pKa = 3.73KK528 pKa = 11.32LVLGTNAPSAISNEE542 pKa = 3.77KK543 pKa = 9.97SGLVIDD549 pKa = 4.28VGTNAGITITATQPTTVVTDD569 pKa = 3.75KK570 pKa = 11.06LATGTVTVDD579 pKa = 3.99GYY581 pKa = 9.63STAAITIDD589 pKa = 3.62GVSITPVGYY598 pKa = 9.24TDD600 pKa = 6.04AIVTKK605 pKa = 9.44TDD607 pKa = 3.45YY608 pKa = 9.88YY609 pKa = 10.89TLEE612 pKa = 5.11LDD614 pKa = 5.02AGDD617 pKa = 3.79TTATVAIEE625 pKa = 3.95YY626 pKa = 10.43DD627 pKa = 3.63SAITEE632 pKa = 4.19LTLVDD637 pKa = 4.04TNLEE641 pKa = 4.46AISNANTATVINVGSISGVEE661 pKa = 3.46ITAAVDD667 pKa = 3.43TTVVTTEE674 pKa = 4.38LSDD677 pKa = 3.72SNTVVLIGDD686 pKa = 3.6KK687 pKa = 10.76TFTLLVLDD695 pKa = 4.67DD696 pKa = 5.18GGYY699 pKa = 10.86EE700 pKa = 4.49DD701 pKa = 6.13DD702 pKa = 4.3PTPGTVTGSTNGYY715 pKa = 9.79VYY717 pKa = 10.38TYY719 pKa = 11.15LNDD722 pKa = 3.58NGNEE726 pKa = 4.0NTEE729 pKa = 4.13FAPIDD734 pKa = 4.19DD735 pKa = 5.37DD736 pKa = 5.11SFSLTSDD743 pKa = 3.02SGTVTVTVSDD753 pKa = 3.53ITPP756 pKa = 3.46
Molecular weight: 78.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F5YLI7|F5YLI7_TREPZ Precorrin-2 dehydrogenase OS=Treponema primitia (strain ATCC BAA-887 / DSM 12427 / ZAS-2) OX=545694 GN=TREPR_0259 PE=4 SV=1
MM1 pKa = 7.04KK2 pKa = 10.01TVDD5 pKa = 3.27PVQAYY10 pKa = 8.89RR11 pKa = 11.84AIRR14 pKa = 11.84KK15 pKa = 9.24RR16 pKa = 11.84CFLLGLLLWVALVLAVIFTLGMGAMDD42 pKa = 4.48IPASSILRR50 pKa = 11.84ILSAKK55 pKa = 9.93LRR57 pKa = 11.84TAFFGPVGSPEE68 pKa = 3.87ALAGISSGMVAVVWEE83 pKa = 4.14LRR85 pKa = 11.84LPRR88 pKa = 11.84ILLSILAGAGLAVAGVIFQGILQNPLADD116 pKa = 4.89PYY118 pKa = 11.1TLGISTGAAFGASLAIFFNITLGLLLPVSSAALVFAALTLALVLLIAQRR167 pKa = 11.84SSGLVSANLIMAGIILSAILSAGISFLKK195 pKa = 9.83MLSGEE200 pKa = 4.19NVGAIVFWLMGSLSAKK216 pKa = 9.21GWNEE220 pKa = 3.5ALLLATVVPAALVLARR236 pKa = 11.84IFAADD241 pKa = 4.23LNILTLGSRR250 pKa = 11.84SAEE253 pKa = 4.0SLGVQVKK260 pKa = 7.83RR261 pKa = 11.84TRR263 pKa = 11.84LFYY266 pKa = 11.0LFLGATISAVCVSTCGVIGFVGLIVPHH293 pKa = 6.74LLRR296 pKa = 11.84MAVTSDD302 pKa = 2.91NRR304 pKa = 11.84LLIPLSALLGGLLLCAADD322 pKa = 3.7TFARR326 pKa = 11.84LLSGGEE332 pKa = 3.88IPVGVLTTLLGGPFFIFIFLRR353 pKa = 11.84QKK355 pKa = 10.58GGRR358 pKa = 11.84GLL360 pKa = 3.58
MM1 pKa = 7.04KK2 pKa = 10.01TVDD5 pKa = 3.27PVQAYY10 pKa = 8.89RR11 pKa = 11.84AIRR14 pKa = 11.84KK15 pKa = 9.24RR16 pKa = 11.84CFLLGLLLWVALVLAVIFTLGMGAMDD42 pKa = 4.48IPASSILRR50 pKa = 11.84ILSAKK55 pKa = 9.93LRR57 pKa = 11.84TAFFGPVGSPEE68 pKa = 3.87ALAGISSGMVAVVWEE83 pKa = 4.14LRR85 pKa = 11.84LPRR88 pKa = 11.84ILLSILAGAGLAVAGVIFQGILQNPLADD116 pKa = 4.89PYY118 pKa = 11.1TLGISTGAAFGASLAIFFNITLGLLLPVSSAALVFAALTLALVLLIAQRR167 pKa = 11.84SSGLVSANLIMAGIILSAILSAGISFLKK195 pKa = 9.83MLSGEE200 pKa = 4.19NVGAIVFWLMGSLSAKK216 pKa = 9.21GWNEE220 pKa = 3.5ALLLATVVPAALVLARR236 pKa = 11.84IFAADD241 pKa = 4.23LNILTLGSRR250 pKa = 11.84SAEE253 pKa = 4.0SLGVQVKK260 pKa = 7.83RR261 pKa = 11.84TRR263 pKa = 11.84LFYY266 pKa = 11.0LFLGATISAVCVSTCGVIGFVGLIVPHH293 pKa = 6.74LLRR296 pKa = 11.84MAVTSDD302 pKa = 2.91NRR304 pKa = 11.84LLIPLSALLGGLLLCAADD322 pKa = 3.7TFARR326 pKa = 11.84LLSGGEE332 pKa = 3.88IPVGVLTTLLGGPFFIFIFLRR353 pKa = 11.84QKK355 pKa = 10.58GGRR358 pKa = 11.84GLL360 pKa = 3.58
Molecular weight: 37.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1208744 |
30 |
6174 |
344.2 |
38.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.727 ± 0.052 | 0.989 ± 0.018 |
5.433 ± 0.035 | 6.495 ± 0.049 |
4.497 ± 0.033 | 7.972 ± 0.048 |
1.533 ± 0.018 | 7.241 ± 0.04 |
5.52 ± 0.051 | 9.84 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.255 ± 0.023 | 4.091 ± 0.032 |
4.506 ± 0.032 | 2.962 ± 0.023 |
5.277 ± 0.044 | 6.345 ± 0.039 |
5.489 ± 0.072 | 6.298 ± 0.039 |
1.063 ± 0.016 | 3.466 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
