
Beihai sobemo-like virus 16
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 5.94
Get precalculated fractions of proteins
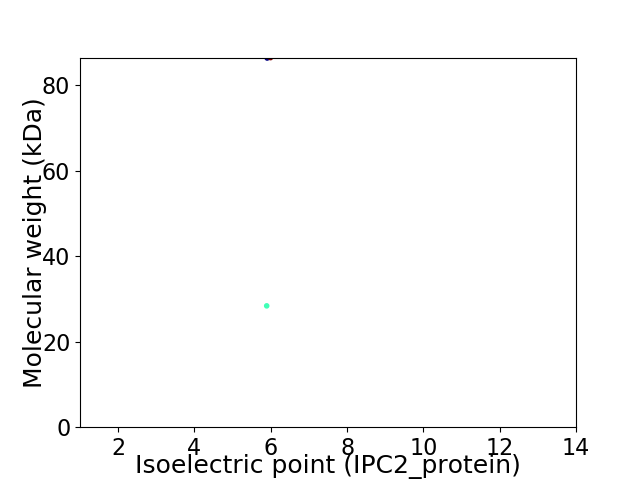
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KE58|A0A1L3KE58_9VIRU RdRP_1 domain-containing protein OS=Beihai sobemo-like virus 16 OX=1922687 PE=4 SV=1
MM1 pKa = 7.4FRR3 pKa = 11.84PIMRR7 pKa = 11.84SVQADD12 pKa = 3.75CLSNGIAIGWAPNLPGGLRR31 pKa = 11.84TLWSHH36 pKa = 6.5IRR38 pKa = 11.84NKK40 pKa = 10.35DD41 pKa = 3.03LCTDD45 pKa = 3.4KK46 pKa = 11.43SSFDD50 pKa = 3.36WTVQPWMMEE59 pKa = 3.88FVAEE63 pKa = 4.42LFSSLVDD70 pKa = 3.61PANSTVVYY78 pKa = 8.16NHH80 pKa = 6.71CCSMLGAKK88 pKa = 8.64TFTAPGVSFHH98 pKa = 6.9HH99 pKa = 6.49PHH101 pKa = 7.42DD102 pKa = 4.41GVLPSGWKK110 pKa = 8.39LTILVNSILQLSFHH124 pKa = 6.62EE125 pKa = 4.17LAGGIGSIIAMGDD138 pKa = 3.18DD139 pKa = 4.53TIQDD143 pKa = 3.46RR144 pKa = 11.84VDD146 pKa = 3.16NTPAYY151 pKa = 9.56MEE153 pKa = 4.44ALSRR157 pKa = 11.84LGPLIKK163 pKa = 10.39VATIGHH169 pKa = 6.51EE170 pKa = 4.06FCGMEE175 pKa = 4.26FTAEE179 pKa = 4.0KK180 pKa = 10.13YY181 pKa = 10.85LPVYY185 pKa = 10.18LEE187 pKa = 3.83KK188 pKa = 10.6HH189 pKa = 5.91SYY191 pKa = 9.82ALRR194 pKa = 11.84HH195 pKa = 5.84LKK197 pKa = 10.74ASVAVEE203 pKa = 3.99TLASYY208 pKa = 10.64QYY210 pKa = 11.37LYY212 pKa = 10.79AYY214 pKa = 10.62DD215 pKa = 3.96NARR218 pKa = 11.84LAAIQDD224 pKa = 3.3WMRR227 pKa = 11.84QLGSPDD233 pKa = 3.21LVKK236 pKa = 10.59PRR238 pKa = 11.84LQLQFEE244 pKa = 4.31PRR246 pKa = 11.84GWSIPISYY254 pKa = 10.56
MM1 pKa = 7.4FRR3 pKa = 11.84PIMRR7 pKa = 11.84SVQADD12 pKa = 3.75CLSNGIAIGWAPNLPGGLRR31 pKa = 11.84TLWSHH36 pKa = 6.5IRR38 pKa = 11.84NKK40 pKa = 10.35DD41 pKa = 3.03LCTDD45 pKa = 3.4KK46 pKa = 11.43SSFDD50 pKa = 3.36WTVQPWMMEE59 pKa = 3.88FVAEE63 pKa = 4.42LFSSLVDD70 pKa = 3.61PANSTVVYY78 pKa = 8.16NHH80 pKa = 6.71CCSMLGAKK88 pKa = 8.64TFTAPGVSFHH98 pKa = 6.9HH99 pKa = 6.49PHH101 pKa = 7.42DD102 pKa = 4.41GVLPSGWKK110 pKa = 8.39LTILVNSILQLSFHH124 pKa = 6.62EE125 pKa = 4.17LAGGIGSIIAMGDD138 pKa = 3.18DD139 pKa = 4.53TIQDD143 pKa = 3.46RR144 pKa = 11.84VDD146 pKa = 3.16NTPAYY151 pKa = 9.56MEE153 pKa = 4.44ALSRR157 pKa = 11.84LGPLIKK163 pKa = 10.39VATIGHH169 pKa = 6.51EE170 pKa = 4.06FCGMEE175 pKa = 4.26FTAEE179 pKa = 4.0KK180 pKa = 10.13YY181 pKa = 10.85LPVYY185 pKa = 10.18LEE187 pKa = 3.83KK188 pKa = 10.6HH189 pKa = 5.91SYY191 pKa = 9.82ALRR194 pKa = 11.84HH195 pKa = 5.84LKK197 pKa = 10.74ASVAVEE203 pKa = 3.99TLASYY208 pKa = 10.64QYY210 pKa = 11.37LYY212 pKa = 10.79AYY214 pKa = 10.62DD215 pKa = 3.96NARR218 pKa = 11.84LAAIQDD224 pKa = 3.3WMRR227 pKa = 11.84QLGSPDD233 pKa = 3.21LVKK236 pKa = 10.59PRR238 pKa = 11.84LQLQFEE244 pKa = 4.31PRR246 pKa = 11.84GWSIPISYY254 pKa = 10.56
Molecular weight: 28.39 kDa
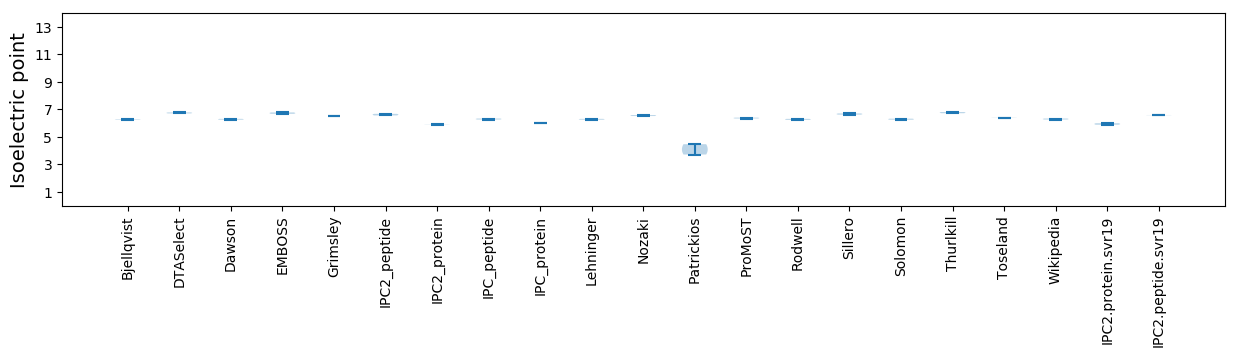
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KE58|A0A1L3KE58_9VIRU RdRP_1 domain-containing protein OS=Beihai sobemo-like virus 16 OX=1922687 PE=4 SV=1
MM1 pKa = 7.45QLFTLSLGLTFSLWLLTRR19 pKa = 11.84TAAATLQNEE28 pKa = 4.54RR29 pKa = 11.84FYY31 pKa = 11.57SSTDD35 pKa = 3.19SFDD38 pKa = 3.32VDD40 pKa = 3.76PVMVAYY46 pKa = 8.26FHH48 pKa = 6.45YY49 pKa = 10.81AFLFFFLVTMIGMTVCFILCPIFLWISRR77 pKa = 11.84TIGYY81 pKa = 9.29CLLGTYY87 pKa = 10.55LPGFKK92 pKa = 10.61LEE94 pKa = 4.75LLNEE98 pKa = 3.85MSVRR102 pKa = 11.84EE103 pKa = 4.23KK104 pKa = 10.21KK105 pKa = 9.13TDD107 pKa = 4.26FIVGAPLPDD116 pKa = 3.69CQVIVYY122 pKa = 10.0RR123 pKa = 11.84RR124 pKa = 11.84LNDD127 pKa = 3.79DD128 pKa = 3.34YY129 pKa = 11.08VTAVGQGFFVRR140 pKa = 11.84DD141 pKa = 3.83GEE143 pKa = 4.74IVTAQHH149 pKa = 5.74NVEE152 pKa = 4.38TAGEE156 pKa = 4.38YY157 pKa = 10.35YY158 pKa = 9.97VAKK161 pKa = 10.47GHH163 pKa = 6.19TEE165 pKa = 4.02LKK167 pKa = 10.64FKK169 pKa = 10.14VTGDD173 pKa = 3.65RR174 pKa = 11.84EE175 pKa = 4.39LEE177 pKa = 4.32GPQIDD182 pKa = 4.34LVVLQVPKK190 pKa = 11.02GVMQKK195 pKa = 8.04MHH197 pKa = 7.42ASPAKK202 pKa = 9.66LQSRR206 pKa = 11.84VLNNQGITIHH216 pKa = 6.89AGGLRR221 pKa = 11.84DD222 pKa = 4.45DD223 pKa = 4.39DD224 pKa = 4.45TVLSSNGTIAAMSGRR239 pKa = 11.84EE240 pKa = 3.9GVLTCGHH247 pKa = 6.4TADD250 pKa = 4.42TLPGFSGAPIFLGRR264 pKa = 11.84KK265 pKa = 7.92VVAIHH270 pKa = 5.49MTSIPSKK277 pKa = 9.64VNSGSWIWPALGDD290 pKa = 3.67YY291 pKa = 7.45FHH293 pKa = 7.56PVLSRR298 pKa = 11.84RR299 pKa = 11.84EE300 pKa = 3.75TAAFSSSTSRR310 pKa = 11.84KK311 pKa = 10.06SSDD314 pKa = 2.27IDD316 pKa = 3.22QFMEE320 pKa = 3.95YY321 pKa = 10.7DD322 pKa = 3.34EE323 pKa = 5.31DD324 pKa = 4.49VYY326 pKa = 11.06IRR328 pKa = 11.84SRR330 pKa = 11.84SGQLRR335 pKa = 11.84KK336 pKa = 9.3MKK338 pKa = 10.74AEE340 pKa = 4.21DD341 pKa = 3.47FFEE344 pKa = 4.43YY345 pKa = 10.88AEE347 pKa = 4.16PTFLRR352 pKa = 11.84SYY354 pKa = 10.86EE355 pKa = 4.13EE356 pKa = 3.89NQEE359 pKa = 4.17LLEE362 pKa = 4.45QGASWAEE369 pKa = 3.69VSKK372 pKa = 10.31WADD375 pKa = 3.38EE376 pKa = 4.25DD377 pKa = 3.59QLAQRR382 pKa = 11.84EE383 pKa = 4.55TNKK386 pKa = 9.73TPVSDD391 pKa = 3.98TDD393 pKa = 4.09SDD395 pKa = 4.74DD396 pKa = 4.02SDD398 pKa = 3.87STSSFVSTKK407 pKa = 10.37SSSSSYY413 pKa = 10.33ASCRR417 pKa = 11.84PEE419 pKa = 3.58PLARR423 pKa = 11.84RR424 pKa = 11.84EE425 pKa = 3.99NLDD428 pKa = 4.56PIPLPTEE435 pKa = 3.7NLRR438 pKa = 11.84PQAVLDD444 pKa = 3.88AVTEE448 pKa = 3.9YY449 pKa = 11.15AIILRR454 pKa = 11.84RR455 pKa = 11.84YY456 pKa = 9.47NEE458 pKa = 4.47LKK460 pKa = 11.16VMIDD464 pKa = 3.47ILNHH468 pKa = 6.27ASNHH472 pKa = 5.18SLALRR477 pKa = 11.84DD478 pKa = 3.69QQSNFQLSSDD488 pKa = 2.99IDD490 pKa = 3.54VLRR493 pKa = 11.84RR494 pKa = 11.84EE495 pKa = 4.72LEE497 pKa = 3.92NASAVLKK504 pKa = 10.49EE505 pKa = 4.37SNLKK509 pKa = 10.3LSHH512 pKa = 6.73IMQKK516 pKa = 10.27SADD519 pKa = 3.92DD520 pKa = 3.84AQVARR525 pKa = 11.84NWQRR529 pKa = 11.84YY530 pKa = 5.18RR531 pKa = 11.84HH532 pKa = 5.1SHH534 pKa = 6.18ADD536 pKa = 3.01VRR538 pKa = 11.84PPVSQQTQDD547 pKa = 3.4LLAAARR553 pKa = 11.84MEE555 pKa = 4.16IDD557 pKa = 3.38AASQRR562 pKa = 11.84LQAIPEE568 pKa = 4.14RR569 pKa = 11.84PRR571 pKa = 11.84QEE573 pKa = 3.65PFAVRR578 pKa = 11.84EE579 pKa = 4.24SVEE582 pKa = 3.7ADD584 pKa = 3.17AVISRR589 pKa = 11.84YY590 pKa = 10.14LPGFFNRR597 pKa = 11.84GAHH600 pKa = 5.98PAGPSVSKK608 pKa = 10.48PSNQTIQAFLPEE620 pKa = 4.75KK621 pKa = 10.51DD622 pKa = 4.12VNPQGNKK629 pKa = 9.98LSSAQPNGSDD639 pKa = 3.53KK640 pKa = 11.15APPPTPDD647 pKa = 4.8DD648 pKa = 4.08SLLLPPPLKK657 pKa = 10.66DD658 pKa = 3.21SSPPTTPIRR667 pKa = 11.84STTVAPTTPSTAAKK681 pKa = 9.88SSPKK685 pKa = 9.48RR686 pKa = 11.84RR687 pKa = 11.84RR688 pKa = 11.84RR689 pKa = 11.84SRR691 pKa = 11.84NSSRR695 pKa = 11.84RR696 pKa = 11.84RR697 pKa = 11.84KK698 pKa = 6.19TTSNPQNPSTPQPSAPHH715 pKa = 5.7SASTRR720 pKa = 11.84EE721 pKa = 4.07SLVPPGVDD729 pKa = 2.52ATLQIEE735 pKa = 4.52KK736 pKa = 10.66LSLAQRR742 pKa = 11.84KK743 pKa = 9.0SLMKK747 pKa = 10.59DD748 pKa = 3.35LLRR751 pKa = 11.84DD752 pKa = 3.28PDD754 pKa = 3.83AYY756 pKa = 11.35LLVGPGEE763 pKa = 4.55NSPSSPPSSPSEE775 pKa = 4.01VSSPPLL781 pKa = 3.55
MM1 pKa = 7.45QLFTLSLGLTFSLWLLTRR19 pKa = 11.84TAAATLQNEE28 pKa = 4.54RR29 pKa = 11.84FYY31 pKa = 11.57SSTDD35 pKa = 3.19SFDD38 pKa = 3.32VDD40 pKa = 3.76PVMVAYY46 pKa = 8.26FHH48 pKa = 6.45YY49 pKa = 10.81AFLFFFLVTMIGMTVCFILCPIFLWISRR77 pKa = 11.84TIGYY81 pKa = 9.29CLLGTYY87 pKa = 10.55LPGFKK92 pKa = 10.61LEE94 pKa = 4.75LLNEE98 pKa = 3.85MSVRR102 pKa = 11.84EE103 pKa = 4.23KK104 pKa = 10.21KK105 pKa = 9.13TDD107 pKa = 4.26FIVGAPLPDD116 pKa = 3.69CQVIVYY122 pKa = 10.0RR123 pKa = 11.84RR124 pKa = 11.84LNDD127 pKa = 3.79DD128 pKa = 3.34YY129 pKa = 11.08VTAVGQGFFVRR140 pKa = 11.84DD141 pKa = 3.83GEE143 pKa = 4.74IVTAQHH149 pKa = 5.74NVEE152 pKa = 4.38TAGEE156 pKa = 4.38YY157 pKa = 10.35YY158 pKa = 9.97VAKK161 pKa = 10.47GHH163 pKa = 6.19TEE165 pKa = 4.02LKK167 pKa = 10.64FKK169 pKa = 10.14VTGDD173 pKa = 3.65RR174 pKa = 11.84EE175 pKa = 4.39LEE177 pKa = 4.32GPQIDD182 pKa = 4.34LVVLQVPKK190 pKa = 11.02GVMQKK195 pKa = 8.04MHH197 pKa = 7.42ASPAKK202 pKa = 9.66LQSRR206 pKa = 11.84VLNNQGITIHH216 pKa = 6.89AGGLRR221 pKa = 11.84DD222 pKa = 4.45DD223 pKa = 4.39DD224 pKa = 4.45TVLSSNGTIAAMSGRR239 pKa = 11.84EE240 pKa = 3.9GVLTCGHH247 pKa = 6.4TADD250 pKa = 4.42TLPGFSGAPIFLGRR264 pKa = 11.84KK265 pKa = 7.92VVAIHH270 pKa = 5.49MTSIPSKK277 pKa = 9.64VNSGSWIWPALGDD290 pKa = 3.67YY291 pKa = 7.45FHH293 pKa = 7.56PVLSRR298 pKa = 11.84RR299 pKa = 11.84EE300 pKa = 3.75TAAFSSSTSRR310 pKa = 11.84KK311 pKa = 10.06SSDD314 pKa = 2.27IDD316 pKa = 3.22QFMEE320 pKa = 3.95YY321 pKa = 10.7DD322 pKa = 3.34EE323 pKa = 5.31DD324 pKa = 4.49VYY326 pKa = 11.06IRR328 pKa = 11.84SRR330 pKa = 11.84SGQLRR335 pKa = 11.84KK336 pKa = 9.3MKK338 pKa = 10.74AEE340 pKa = 4.21DD341 pKa = 3.47FFEE344 pKa = 4.43YY345 pKa = 10.88AEE347 pKa = 4.16PTFLRR352 pKa = 11.84SYY354 pKa = 10.86EE355 pKa = 4.13EE356 pKa = 3.89NQEE359 pKa = 4.17LLEE362 pKa = 4.45QGASWAEE369 pKa = 3.69VSKK372 pKa = 10.31WADD375 pKa = 3.38EE376 pKa = 4.25DD377 pKa = 3.59QLAQRR382 pKa = 11.84EE383 pKa = 4.55TNKK386 pKa = 9.73TPVSDD391 pKa = 3.98TDD393 pKa = 4.09SDD395 pKa = 4.74DD396 pKa = 4.02SDD398 pKa = 3.87STSSFVSTKK407 pKa = 10.37SSSSSYY413 pKa = 10.33ASCRR417 pKa = 11.84PEE419 pKa = 3.58PLARR423 pKa = 11.84RR424 pKa = 11.84EE425 pKa = 3.99NLDD428 pKa = 4.56PIPLPTEE435 pKa = 3.7NLRR438 pKa = 11.84PQAVLDD444 pKa = 3.88AVTEE448 pKa = 3.9YY449 pKa = 11.15AIILRR454 pKa = 11.84RR455 pKa = 11.84YY456 pKa = 9.47NEE458 pKa = 4.47LKK460 pKa = 11.16VMIDD464 pKa = 3.47ILNHH468 pKa = 6.27ASNHH472 pKa = 5.18SLALRR477 pKa = 11.84DD478 pKa = 3.69QQSNFQLSSDD488 pKa = 2.99IDD490 pKa = 3.54VLRR493 pKa = 11.84RR494 pKa = 11.84EE495 pKa = 4.72LEE497 pKa = 3.92NASAVLKK504 pKa = 10.49EE505 pKa = 4.37SNLKK509 pKa = 10.3LSHH512 pKa = 6.73IMQKK516 pKa = 10.27SADD519 pKa = 3.92DD520 pKa = 3.84AQVARR525 pKa = 11.84NWQRR529 pKa = 11.84YY530 pKa = 5.18RR531 pKa = 11.84HH532 pKa = 5.1SHH534 pKa = 6.18ADD536 pKa = 3.01VRR538 pKa = 11.84PPVSQQTQDD547 pKa = 3.4LLAAARR553 pKa = 11.84MEE555 pKa = 4.16IDD557 pKa = 3.38AASQRR562 pKa = 11.84LQAIPEE568 pKa = 4.14RR569 pKa = 11.84PRR571 pKa = 11.84QEE573 pKa = 3.65PFAVRR578 pKa = 11.84EE579 pKa = 4.24SVEE582 pKa = 3.7ADD584 pKa = 3.17AVISRR589 pKa = 11.84YY590 pKa = 10.14LPGFFNRR597 pKa = 11.84GAHH600 pKa = 5.98PAGPSVSKK608 pKa = 10.48PSNQTIQAFLPEE620 pKa = 4.75KK621 pKa = 10.51DD622 pKa = 4.12VNPQGNKK629 pKa = 9.98LSSAQPNGSDD639 pKa = 3.53KK640 pKa = 11.15APPPTPDD647 pKa = 4.8DD648 pKa = 4.08SLLLPPPLKK657 pKa = 10.66DD658 pKa = 3.21SSPPTTPIRR667 pKa = 11.84STTVAPTTPSTAAKK681 pKa = 9.88SSPKK685 pKa = 9.48RR686 pKa = 11.84RR687 pKa = 11.84RR688 pKa = 11.84RR689 pKa = 11.84SRR691 pKa = 11.84NSSRR695 pKa = 11.84RR696 pKa = 11.84RR697 pKa = 11.84KK698 pKa = 6.19TTSNPQNPSTPQPSAPHH715 pKa = 5.7SASTRR720 pKa = 11.84EE721 pKa = 4.07SLVPPGVDD729 pKa = 2.52ATLQIEE735 pKa = 4.52KK736 pKa = 10.66LSLAQRR742 pKa = 11.84KK743 pKa = 9.0SLMKK747 pKa = 10.59DD748 pKa = 3.35LLRR751 pKa = 11.84DD752 pKa = 3.28PDD754 pKa = 3.83AYY756 pKa = 11.35LLVGPGEE763 pKa = 4.55NSPSSPPSSPSEE775 pKa = 4.01VSSPPLL781 pKa = 3.55
Molecular weight: 86.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1035 |
254 |
781 |
517.5 |
57.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.019 ± 0.115 | 1.063 ± 0.419 |
5.894 ± 0.359 | 5.024 ± 0.503 |
3.865 ± 0.033 | 5.121 ± 0.727 |
2.319 ± 0.567 | 4.444 ± 0.676 |
3.961 ± 0.193 | 9.855 ± 0.541 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.319 ± 0.567 | 3.478 ± 0.152 |
7.343 ± 0.665 | 4.444 ± 0.417 |
6.087 ± 0.813 | 10.531 ± 1.047 |
5.797 ± 0.496 | 6.184 ± 0.129 |
1.353 ± 0.649 | 2.899 ± 0.481 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
