
Mycolicibacter sinensis (strain JDM601) (Mycobacterium sinense)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Mycobacteriaceae; Mycolicibacter
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
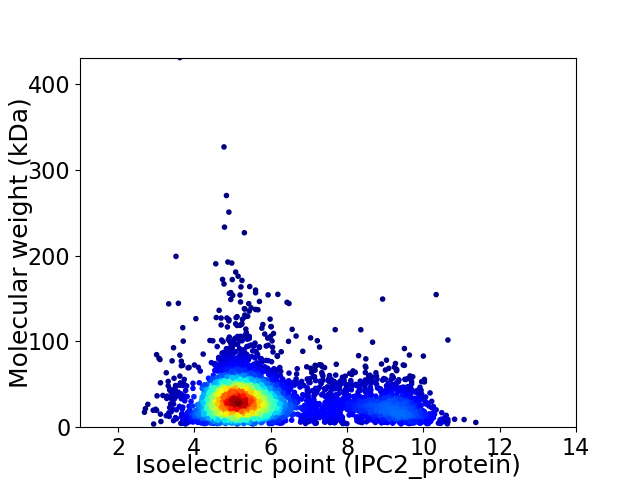
Virtual 2D-PAGE plot for 4332 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F5YVZ4|F5YVZ4_MYCSD MarR family transcriptional regulator OS=Mycolicibacter sinensis (strain JDM601) OX=875328 GN=JDM601_1241 PE=4 SV=1
MM1 pKa = 6.5TTIKK5 pKa = 10.57KK6 pKa = 8.23FLCGATVAGMVAGTGAAVGVPSGLSALTADD36 pKa = 3.29WLLAAEE42 pKa = 4.4HH43 pKa = 6.29VLLIGTDD50 pKa = 3.59GTNLSKK56 pKa = 10.53ILEE59 pKa = 4.17YY60 pKa = 10.56AYY62 pKa = 10.7GDD64 pKa = 3.47DD65 pKa = 4.72SGFKK69 pKa = 9.44TAMDD73 pKa = 3.52QGITGATSEE82 pKa = 4.49VNHH85 pKa = 5.46TTISGPSWSTILTGVWDD102 pKa = 4.53DD103 pKa = 3.32KK104 pKa = 11.56HH105 pKa = 7.68GVVNNLFRR113 pKa = 11.84PEE115 pKa = 5.49PYY117 pKa = 10.21DD118 pKa = 3.07QWPTVFNLLEE128 pKa = 3.99YY129 pKa = 10.26HH130 pKa = 6.39NPDD133 pKa = 3.08IEE135 pKa = 4.45TSVIADD141 pKa = 3.24WDD143 pKa = 4.2YY144 pKa = 11.77INDD147 pKa = 3.29IGAAGGYY154 pKa = 9.71GADD157 pKa = 3.63NNLFIPFQDD166 pKa = 2.99SWADD170 pKa = 2.94TDD172 pKa = 4.13AAVTAATISQILATTTNPDD191 pKa = 2.41IDD193 pKa = 4.0TFLFSYY199 pKa = 9.78QVQVDD204 pKa = 3.97EE205 pKa = 5.33AGHH208 pKa = 5.71SFGGGSQEE216 pKa = 3.92YY217 pKa = 9.29AQAVMNVGANIQQILAAVEE236 pKa = 3.89QARR239 pKa = 11.84QEE241 pKa = 4.34TGDD244 pKa = 3.31NWSIIITTDD253 pKa = 2.75HH254 pKa = 6.28GHH256 pKa = 4.9QQSLGFGHH264 pKa = 7.34GFQSPNEE271 pKa = 3.97TSQFVIFDD279 pKa = 3.88QAGHH283 pKa = 7.24DD284 pKa = 3.87ATDD287 pKa = 4.02GSQNLNYY294 pKa = 10.09SIADD298 pKa = 3.31ITPTILQLLGVPLRR312 pKa = 11.84SDD314 pKa = 3.79FDD316 pKa = 4.14GSPMSTDD323 pKa = 3.46PAILDD328 pKa = 4.16SIVTPTDD335 pKa = 3.64LKK337 pKa = 11.09QSLSDD342 pKa = 4.46AIAMYY347 pKa = 10.04GYY349 pKa = 9.73PNIGNDD355 pKa = 2.55IMLGIRR361 pKa = 11.84TVVTSIPYY369 pKa = 10.07GINLAVTEE377 pKa = 4.26IDD379 pKa = 4.45KK380 pKa = 10.68FLQSIVDD387 pKa = 3.35QDD389 pKa = 3.21IFLISGLAEE398 pKa = 4.36LLQQVNNFVGGAVVDD413 pKa = 4.14VTDD416 pKa = 3.57TLARR420 pKa = 11.84GVAYY424 pKa = 8.73LTGSGVIAPTDD435 pKa = 3.8PPLPPPPGGSEE446 pKa = 3.79ASLFDD451 pKa = 3.99TLLSDD456 pKa = 5.09HH457 pKa = 6.68SWLSPDD463 pKa = 3.87EE464 pKa = 4.43GSFDD468 pKa = 4.34LASLLDD474 pKa = 4.81FSALLDD480 pKa = 3.63
MM1 pKa = 6.5TTIKK5 pKa = 10.57KK6 pKa = 8.23FLCGATVAGMVAGTGAAVGVPSGLSALTADD36 pKa = 3.29WLLAAEE42 pKa = 4.4HH43 pKa = 6.29VLLIGTDD50 pKa = 3.59GTNLSKK56 pKa = 10.53ILEE59 pKa = 4.17YY60 pKa = 10.56AYY62 pKa = 10.7GDD64 pKa = 3.47DD65 pKa = 4.72SGFKK69 pKa = 9.44TAMDD73 pKa = 3.52QGITGATSEE82 pKa = 4.49VNHH85 pKa = 5.46TTISGPSWSTILTGVWDD102 pKa = 4.53DD103 pKa = 3.32KK104 pKa = 11.56HH105 pKa = 7.68GVVNNLFRR113 pKa = 11.84PEE115 pKa = 5.49PYY117 pKa = 10.21DD118 pKa = 3.07QWPTVFNLLEE128 pKa = 3.99YY129 pKa = 10.26HH130 pKa = 6.39NPDD133 pKa = 3.08IEE135 pKa = 4.45TSVIADD141 pKa = 3.24WDD143 pKa = 4.2YY144 pKa = 11.77INDD147 pKa = 3.29IGAAGGYY154 pKa = 9.71GADD157 pKa = 3.63NNLFIPFQDD166 pKa = 2.99SWADD170 pKa = 2.94TDD172 pKa = 4.13AAVTAATISQILATTTNPDD191 pKa = 2.41IDD193 pKa = 4.0TFLFSYY199 pKa = 9.78QVQVDD204 pKa = 3.97EE205 pKa = 5.33AGHH208 pKa = 5.71SFGGGSQEE216 pKa = 3.92YY217 pKa = 9.29AQAVMNVGANIQQILAAVEE236 pKa = 3.89QARR239 pKa = 11.84QEE241 pKa = 4.34TGDD244 pKa = 3.31NWSIIITTDD253 pKa = 2.75HH254 pKa = 6.28GHH256 pKa = 4.9QQSLGFGHH264 pKa = 7.34GFQSPNEE271 pKa = 3.97TSQFVIFDD279 pKa = 3.88QAGHH283 pKa = 7.24DD284 pKa = 3.87ATDD287 pKa = 4.02GSQNLNYY294 pKa = 10.09SIADD298 pKa = 3.31ITPTILQLLGVPLRR312 pKa = 11.84SDD314 pKa = 3.79FDD316 pKa = 4.14GSPMSTDD323 pKa = 3.46PAILDD328 pKa = 4.16SIVTPTDD335 pKa = 3.64LKK337 pKa = 11.09QSLSDD342 pKa = 4.46AIAMYY347 pKa = 10.04GYY349 pKa = 9.73PNIGNDD355 pKa = 2.55IMLGIRR361 pKa = 11.84TVVTSIPYY369 pKa = 10.07GINLAVTEE377 pKa = 4.26IDD379 pKa = 4.45KK380 pKa = 10.68FLQSIVDD387 pKa = 3.35QDD389 pKa = 3.21IFLISGLAEE398 pKa = 4.36LLQQVNNFVGGAVVDD413 pKa = 4.14VTDD416 pKa = 3.57TLARR420 pKa = 11.84GVAYY424 pKa = 8.73LTGSGVIAPTDD435 pKa = 3.8PPLPPPPGGSEE446 pKa = 3.79ASLFDD451 pKa = 3.99TLLSDD456 pKa = 5.09HH457 pKa = 6.68SWLSPDD463 pKa = 3.87EE464 pKa = 4.43GSFDD468 pKa = 4.34LASLLDD474 pKa = 4.81FSALLDD480 pKa = 3.63
Molecular weight: 50.89 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F5YWB2|F5YWB2_MYCSD Uncharacterized protein OS=Mycolicibacter sinensis (strain JDM601) OX=875328 GN=JDM601_0013 PE=3 SV=1
MM1 pKa = 7.69AKK3 pKa = 10.06GKK5 pKa = 8.69RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84VHH19 pKa = 5.99GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AIVTGRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.68GRR42 pKa = 11.84RR43 pKa = 11.84SLTAA47 pKa = 3.9
MM1 pKa = 7.69AKK3 pKa = 10.06GKK5 pKa = 8.69RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84VHH19 pKa = 5.99GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AIVTGRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.68GRR42 pKa = 11.84RR43 pKa = 11.84SLTAA47 pKa = 3.9
Molecular weight: 5.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1398531 |
37 |
5188 |
322.8 |
34.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.601 ± 0.051 | 0.794 ± 0.01 |
6.302 ± 0.035 | 5.145 ± 0.033 |
2.989 ± 0.027 | 9.656 ± 0.158 |
2.192 ± 0.018 | 4.103 ± 0.027 |
2.017 ± 0.025 | 9.787 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.037 ± 0.019 | 2.229 ± 0.024 |
5.822 ± 0.043 | 3.043 ± 0.018 |
7.165 ± 0.048 | 5.175 ± 0.026 |
5.952 ± 0.037 | 8.389 ± 0.038 |
1.479 ± 0.016 | 2.123 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
