
Mesonia phycicola
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Mesonia
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
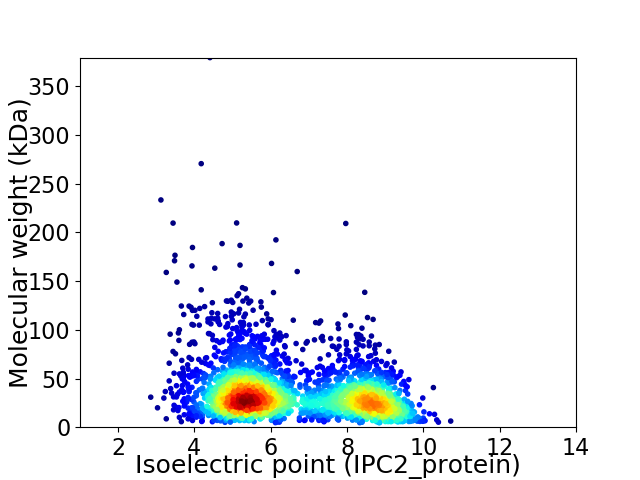
Virtual 2D-PAGE plot for 2974 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M6C3A9|A0A1M6C3A9_9FLAO Glucoamylase (Glucan-1 4-alpha-glucosidase) GH15 family OS=Mesonia phycicola OX=579105 GN=SAMN04488096_102334 PE=4 SV=1
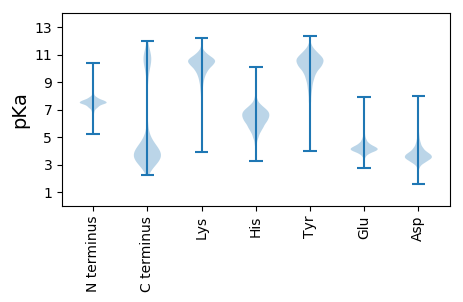
MM1 pKa = 7.58IKK3 pKa = 10.31GKK5 pKa = 10.57KK6 pKa = 9.65FFIGLALASLGVACSSDD23 pKa = 3.65DD24 pKa = 4.63DD25 pKa = 4.29FDD27 pKa = 4.7RR28 pKa = 11.84GNWVEE33 pKa = 4.23SSVFDD38 pKa = 3.7GVPRR42 pKa = 11.84SSAAAFTIDD51 pKa = 2.93NLGYY55 pKa = 9.95MGTGYY60 pKa = 10.89DD61 pKa = 3.7GDD63 pKa = 5.42DD64 pKa = 3.64YY65 pKa = 11.8LSDD68 pKa = 3.12FWEE71 pKa = 4.23YY72 pKa = 11.1NIEE75 pKa = 3.67GDD77 pKa = 3.85YY78 pKa = 10.02WVQKK82 pKa = 11.07ADD84 pKa = 3.89FPSTPRR90 pKa = 11.84SAASAFTIDD99 pKa = 3.09GKK101 pKa = 11.43GYY103 pKa = 10.56LGLGYY108 pKa = 10.68DD109 pKa = 5.32GDD111 pKa = 6.11DD112 pKa = 3.49EE113 pKa = 5.93LQDD116 pKa = 3.4FWEE119 pKa = 4.56YY120 pKa = 11.59NPATNTWDD128 pKa = 3.62SIADD132 pKa = 3.48FGGGVRR138 pKa = 11.84RR139 pKa = 11.84SATGFAINGTGYY151 pKa = 10.91VGTGYY156 pKa = 10.91DD157 pKa = 3.99GDD159 pKa = 4.53NDD161 pKa = 5.42LKK163 pKa = 11.27DD164 pKa = 3.62FWKK167 pKa = 10.8YY168 pKa = 10.95NPSTDD173 pKa = 2.43QWTEE177 pKa = 3.55MVGFGGGKK185 pKa = 9.59RR186 pKa = 11.84RR187 pKa = 11.84DD188 pKa = 3.16AAYY191 pKa = 10.04FILNDD196 pKa = 3.68LVYY199 pKa = 10.57FGTGVTNGVYY209 pKa = 8.74NTDD212 pKa = 3.67FWQFDD217 pKa = 3.68PSSEE221 pKa = 3.68VWTRR225 pKa = 11.84LNDD228 pKa = 3.7LDD230 pKa = 5.25EE231 pKa = 4.72EE232 pKa = 4.5DD233 pKa = 4.05SYY235 pKa = 12.21SVVRR239 pKa = 11.84SNAVGFSQAGYY250 pKa = 10.15GYY252 pKa = 10.37FATGYY257 pKa = 10.55NGGALDD263 pKa = 4.89TIWEE267 pKa = 4.14YY268 pKa = 11.71DD269 pKa = 3.49PTNDD273 pKa = 3.16DD274 pKa = 3.41WEE276 pKa = 5.22KK277 pKa = 9.5ITEE280 pKa = 4.24FEE282 pKa = 3.81ATARR286 pKa = 11.84QDD288 pKa = 3.21AVSFSTATRR297 pKa = 11.84AFVLLGRR304 pKa = 11.84TGSLYY309 pKa = 10.84LDD311 pKa = 4.19DD312 pKa = 4.76NYY314 pKa = 11.04EE315 pKa = 3.94VFTQDD320 pKa = 4.3EE321 pKa = 4.5YY322 pKa = 11.78DD323 pKa = 3.83EE324 pKa = 4.73EE325 pKa = 4.56DD326 pKa = 3.13
MM1 pKa = 7.58IKK3 pKa = 10.31GKK5 pKa = 10.57KK6 pKa = 9.65FFIGLALASLGVACSSDD23 pKa = 3.65DD24 pKa = 4.63DD25 pKa = 4.29FDD27 pKa = 4.7RR28 pKa = 11.84GNWVEE33 pKa = 4.23SSVFDD38 pKa = 3.7GVPRR42 pKa = 11.84SSAAAFTIDD51 pKa = 2.93NLGYY55 pKa = 9.95MGTGYY60 pKa = 10.89DD61 pKa = 3.7GDD63 pKa = 5.42DD64 pKa = 3.64YY65 pKa = 11.8LSDD68 pKa = 3.12FWEE71 pKa = 4.23YY72 pKa = 11.1NIEE75 pKa = 3.67GDD77 pKa = 3.85YY78 pKa = 10.02WVQKK82 pKa = 11.07ADD84 pKa = 3.89FPSTPRR90 pKa = 11.84SAASAFTIDD99 pKa = 3.09GKK101 pKa = 11.43GYY103 pKa = 10.56LGLGYY108 pKa = 10.68DD109 pKa = 5.32GDD111 pKa = 6.11DD112 pKa = 3.49EE113 pKa = 5.93LQDD116 pKa = 3.4FWEE119 pKa = 4.56YY120 pKa = 11.59NPATNTWDD128 pKa = 3.62SIADD132 pKa = 3.48FGGGVRR138 pKa = 11.84RR139 pKa = 11.84SATGFAINGTGYY151 pKa = 10.91VGTGYY156 pKa = 10.91DD157 pKa = 3.99GDD159 pKa = 4.53NDD161 pKa = 5.42LKK163 pKa = 11.27DD164 pKa = 3.62FWKK167 pKa = 10.8YY168 pKa = 10.95NPSTDD173 pKa = 2.43QWTEE177 pKa = 3.55MVGFGGGKK185 pKa = 9.59RR186 pKa = 11.84RR187 pKa = 11.84DD188 pKa = 3.16AAYY191 pKa = 10.04FILNDD196 pKa = 3.68LVYY199 pKa = 10.57FGTGVTNGVYY209 pKa = 8.74NTDD212 pKa = 3.67FWQFDD217 pKa = 3.68PSSEE221 pKa = 3.68VWTRR225 pKa = 11.84LNDD228 pKa = 3.7LDD230 pKa = 5.25EE231 pKa = 4.72EE232 pKa = 4.5DD233 pKa = 4.05SYY235 pKa = 12.21SVVRR239 pKa = 11.84SNAVGFSQAGYY250 pKa = 10.15GYY252 pKa = 10.37FATGYY257 pKa = 10.55NGGALDD263 pKa = 4.89TIWEE267 pKa = 4.14YY268 pKa = 11.71DD269 pKa = 3.49PTNDD273 pKa = 3.16DD274 pKa = 3.41WEE276 pKa = 5.22KK277 pKa = 9.5ITEE280 pKa = 4.24FEE282 pKa = 3.81ATARR286 pKa = 11.84QDD288 pKa = 3.21AVSFSTATRR297 pKa = 11.84AFVLLGRR304 pKa = 11.84TGSLYY309 pKa = 10.84LDD311 pKa = 4.19DD312 pKa = 4.76NYY314 pKa = 11.04EE315 pKa = 3.94VFTQDD320 pKa = 4.3EE321 pKa = 4.5YY322 pKa = 11.78DD323 pKa = 3.83EE324 pKa = 4.73EE325 pKa = 4.56DD326 pKa = 3.13
Molecular weight: 36.27 kDa
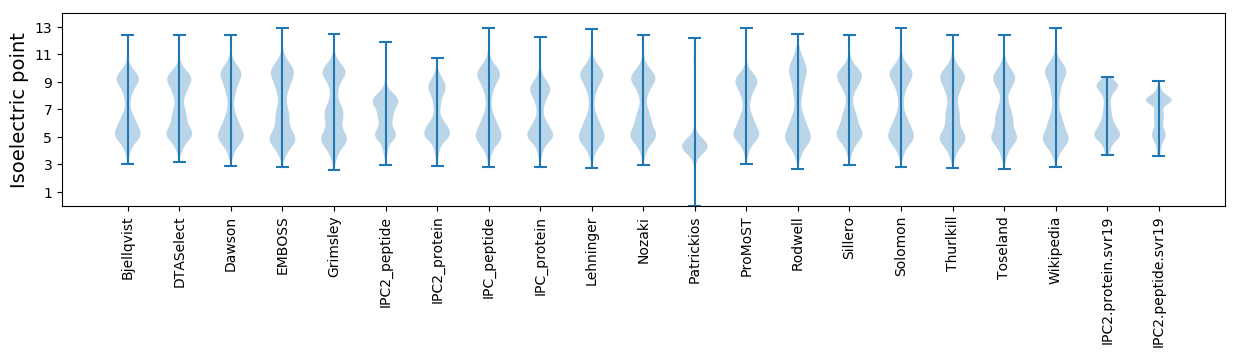
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M6C0F6|A0A1M6C0F6_9FLAO CRP/FNR family transcriptional regulator anaerobic regulatory protein OS=Mesonia phycicola OX=579105 GN=SAMN04488096_102309 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.05GFRR19 pKa = 11.84EE20 pKa = 4.13RR21 pKa = 11.84MSSVNGRR28 pKa = 11.84KK29 pKa = 9.17VLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.14GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.73LSVSSEE47 pKa = 3.76NRR49 pKa = 11.84HH50 pKa = 4.47KK51 pKa = 10.5HH52 pKa = 4.49
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.05GFRR19 pKa = 11.84EE20 pKa = 4.13RR21 pKa = 11.84MSSVNGRR28 pKa = 11.84KK29 pKa = 9.17VLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.14GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.73LSVSSEE47 pKa = 3.76NRR49 pKa = 11.84HH50 pKa = 4.47KK51 pKa = 10.5HH52 pKa = 4.49
Molecular weight: 6.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
992302 |
40 |
3340 |
333.7 |
37.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.095 ± 0.038 | 0.715 ± 0.013 |
5.351 ± 0.042 | 7.202 ± 0.046 |
5.259 ± 0.042 | 5.988 ± 0.046 |
1.692 ± 0.021 | 8.043 ± 0.046 |
8.156 ± 0.071 | 9.391 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.982 ± 0.021 | 6.49 ± 0.047 |
3.204 ± 0.023 | 3.653 ± 0.026 |
3.085 ± 0.028 | 6.568 ± 0.043 |
5.78 ± 0.05 | 6.077 ± 0.034 |
0.998 ± 0.016 | 4.272 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
