
Candidimonas bauzanensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Alcaligenaceae; Candidimonas
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
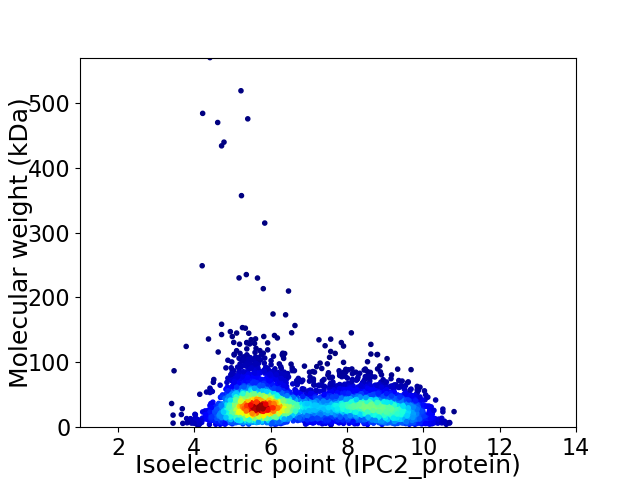
Virtual 2D-PAGE plot for 5118 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M5XCV2|A0A1M5XCV2_9BURK Uncharacterized protein OS=Candidimonas bauzanensis OX=658167 GN=SAMN04488135_106312 PE=4 SV=1
MM1 pKa = 7.85DD2 pKa = 6.75PILATLPPSLLALVEE17 pKa = 4.5GSLSNDD23 pKa = 2.79EE24 pKa = 4.32VSSDD28 pKa = 4.01EE29 pKa = 4.14EE30 pKa = 3.84MLAYY34 pKa = 10.24FIDD37 pKa = 3.95NGLTEE42 pKa = 4.41DD43 pKa = 3.76QARR46 pKa = 11.84QALTYY51 pKa = 10.19RR52 pKa = 11.84DD53 pKa = 3.41QYY55 pKa = 11.36LNNIYY60 pKa = 10.76LEE62 pKa = 4.48GFTPITSADD71 pKa = 3.8EE72 pKa = 4.11PLHH75 pKa = 6.08FNPHH79 pKa = 4.38TRR81 pKa = 11.84QFEE84 pKa = 3.94PDD86 pKa = 2.92
MM1 pKa = 7.85DD2 pKa = 6.75PILATLPPSLLALVEE17 pKa = 4.5GSLSNDD23 pKa = 2.79EE24 pKa = 4.32VSSDD28 pKa = 4.01EE29 pKa = 4.14EE30 pKa = 3.84MLAYY34 pKa = 10.24FIDD37 pKa = 3.95NGLTEE42 pKa = 4.41DD43 pKa = 3.76QARR46 pKa = 11.84QALTYY51 pKa = 10.19RR52 pKa = 11.84DD53 pKa = 3.41QYY55 pKa = 11.36LNNIYY60 pKa = 10.76LEE62 pKa = 4.48GFTPITSADD71 pKa = 3.8EE72 pKa = 4.11PLHH75 pKa = 6.08FNPHH79 pKa = 4.38TRR81 pKa = 11.84QFEE84 pKa = 3.94PDD86 pKa = 2.92
Molecular weight: 9.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M5YQP0|A0A1M5YQP0_9BURK AraC-type DNA-binding protein OS=Candidimonas bauzanensis OX=658167 GN=SAMN04488135_11061 PE=4 SV=1
MM1 pKa = 7.21LQTWLWARR9 pKa = 11.84FKK11 pKa = 10.79GVKK14 pKa = 8.89RR15 pKa = 11.84TVSRR19 pKa = 11.84KK20 pKa = 6.22TRR22 pKa = 11.84QVHH25 pKa = 4.92RR26 pKa = 11.84LSRR29 pKa = 11.84RR30 pKa = 11.84TIQFSLVLVGAALVSFVSLGFAEE53 pKa = 5.04LVDD56 pKa = 4.26LALALNHH63 pKa = 6.74RR64 pKa = 11.84WLQAAPWAVWLILPFGLALLRR85 pKa = 11.84WWTLRR90 pKa = 11.84FAPNAGGSGIPQVIAALSLPADD112 pKa = 3.63SGRR115 pKa = 11.84RR116 pKa = 11.84SLVALSQAAWKK127 pKa = 10.62VPLTFLGLMCGASIGRR143 pKa = 11.84EE144 pKa = 4.25GPSVQVGAALMLAWGQACKK163 pKa = 10.69ALGVPLKK170 pKa = 10.62GIHH173 pKa = 5.21TEE175 pKa = 3.91EE176 pKa = 5.38LIATGAAGGLAAAFNAPLAGVIFAIEE202 pKa = 3.75EE203 pKa = 4.27LGRR206 pKa = 11.84GTALRR211 pKa = 11.84WQRR214 pKa = 11.84LVLIGVLAAGFIVVAFAGNNPYY236 pKa = 10.38FGEE239 pKa = 4.3FTGQALTHH247 pKa = 7.15DD248 pKa = 3.98MLGWTLLCGVACGVGGGLFSWLLSKK273 pKa = 10.79GAASFAPSIWRR284 pKa = 11.84GWIRR288 pKa = 11.84RR289 pKa = 11.84HH290 pKa = 5.2PVWLAFIMGLIVAVLGTLTNNSVYY314 pKa = 9.44GTGYY318 pKa = 10.64GVASTALAGQQLEE331 pKa = 4.51SAGFGPAKK339 pKa = 10.48LLATVASYY347 pKa = 9.18WAGIPGGIFTPALTTGAGIGAEE369 pKa = 4.07LASWVGGVADD379 pKa = 3.78QKK381 pKa = 11.59VLVLLSMAAFLAAATQSPLTASVVVMEE408 pKa = 4.61MTGSQPMLFWLLVCSLVASIVSRR431 pKa = 11.84LFSPLPFYY439 pKa = 10.58HH440 pKa = 6.82HH441 pKa = 6.01SAGRR445 pKa = 11.84FRR447 pKa = 11.84QQVRR451 pKa = 11.84EE452 pKa = 3.95VDD454 pKa = 3.33RR455 pKa = 11.84AQAKK459 pKa = 9.63
MM1 pKa = 7.21LQTWLWARR9 pKa = 11.84FKK11 pKa = 10.79GVKK14 pKa = 8.89RR15 pKa = 11.84TVSRR19 pKa = 11.84KK20 pKa = 6.22TRR22 pKa = 11.84QVHH25 pKa = 4.92RR26 pKa = 11.84LSRR29 pKa = 11.84RR30 pKa = 11.84TIQFSLVLVGAALVSFVSLGFAEE53 pKa = 5.04LVDD56 pKa = 4.26LALALNHH63 pKa = 6.74RR64 pKa = 11.84WLQAAPWAVWLILPFGLALLRR85 pKa = 11.84WWTLRR90 pKa = 11.84FAPNAGGSGIPQVIAALSLPADD112 pKa = 3.63SGRR115 pKa = 11.84RR116 pKa = 11.84SLVALSQAAWKK127 pKa = 10.62VPLTFLGLMCGASIGRR143 pKa = 11.84EE144 pKa = 4.25GPSVQVGAALMLAWGQACKK163 pKa = 10.69ALGVPLKK170 pKa = 10.62GIHH173 pKa = 5.21TEE175 pKa = 3.91EE176 pKa = 5.38LIATGAAGGLAAAFNAPLAGVIFAIEE202 pKa = 3.75EE203 pKa = 4.27LGRR206 pKa = 11.84GTALRR211 pKa = 11.84WQRR214 pKa = 11.84LVLIGVLAAGFIVVAFAGNNPYY236 pKa = 10.38FGEE239 pKa = 4.3FTGQALTHH247 pKa = 7.15DD248 pKa = 3.98MLGWTLLCGVACGVGGGLFSWLLSKK273 pKa = 10.79GAASFAPSIWRR284 pKa = 11.84GWIRR288 pKa = 11.84RR289 pKa = 11.84HH290 pKa = 5.2PVWLAFIMGLIVAVLGTLTNNSVYY314 pKa = 9.44GTGYY318 pKa = 10.64GVASTALAGQQLEE331 pKa = 4.51SAGFGPAKK339 pKa = 10.48LLATVASYY347 pKa = 9.18WAGIPGGIFTPALTTGAGIGAEE369 pKa = 4.07LASWVGGVADD379 pKa = 3.78QKK381 pKa = 11.59VLVLLSMAAFLAAATQSPLTASVVVMEE408 pKa = 4.61MTGSQPMLFWLLVCSLVASIVSRR431 pKa = 11.84LFSPLPFYY439 pKa = 10.58HH440 pKa = 6.82HH441 pKa = 6.01SAGRR445 pKa = 11.84FRR447 pKa = 11.84QQVRR451 pKa = 11.84EE452 pKa = 3.95VDD454 pKa = 3.33RR455 pKa = 11.84AQAKK459 pKa = 9.63
Molecular weight: 48.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1690431 |
39 |
5775 |
330.3 |
35.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.518 ± 0.047 | 0.922 ± 0.012 |
5.264 ± 0.027 | 5.036 ± 0.034 |
3.518 ± 0.022 | 8.523 ± 0.051 |
2.166 ± 0.018 | 5.158 ± 0.027 |
3.285 ± 0.027 | 10.784 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.52 ± 0.019 | 2.777 ± 0.027 |
5.104 ± 0.028 | 3.977 ± 0.021 |
6.676 ± 0.039 | 5.808 ± 0.033 |
4.853 ± 0.034 | 7.21 ± 0.028 |
1.392 ± 0.015 | 2.508 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
