
Carrot mottle mimic virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Calvusvirinae; Umbravirus
Average proteome isoelectric point is 7.59
Get precalculated fractions of proteins
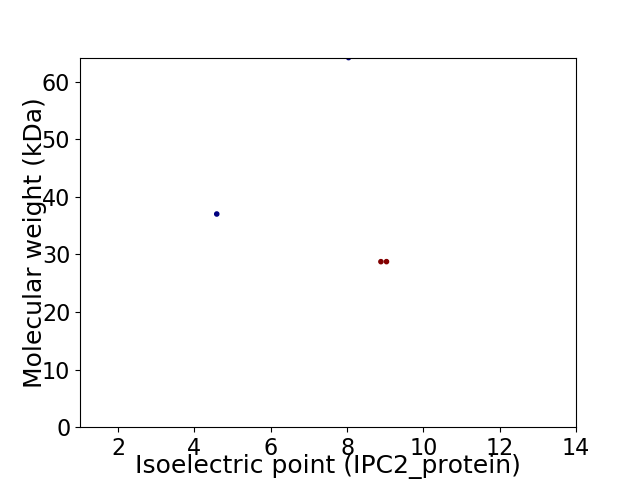
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q66127|Q66127_9TOMB RNA-directed RNA polymerase (Fragment) OS=Carrot mottle mimic virus OX=47736 PE=4 SV=1
MM1 pKa = 7.82DD2 pKa = 6.64HH3 pKa = 5.96ITKK6 pKa = 10.15FLRR9 pKa = 11.84MDD11 pKa = 4.23FSPKK15 pKa = 10.39DD16 pKa = 3.41GVLTRR21 pKa = 11.84RR22 pKa = 11.84QCAEE26 pKa = 4.11RR27 pKa = 11.84FDD29 pKa = 4.32DD30 pKa = 3.7TWGLLITQARR40 pKa = 11.84MTTAHH45 pKa = 6.33TADD48 pKa = 2.91MCAWYY53 pKa = 9.96EE54 pKa = 4.04GSRR57 pKa = 11.84EE58 pKa = 4.08TNQYY62 pKa = 10.81LPLEE66 pKa = 4.22QFVDD70 pKa = 3.45SRR72 pKa = 11.84DD73 pKa = 3.5ASKK76 pKa = 10.6EE77 pKa = 4.02AEE79 pKa = 4.28DD80 pKa = 4.93KK81 pKa = 11.0VDD83 pKa = 4.16KK84 pKa = 10.92LSSPPTRR91 pKa = 11.84GCGGTTVNADD101 pKa = 3.05GGLVEE106 pKa = 5.76NPLPTNRR113 pKa = 11.84DD114 pKa = 3.26EE115 pKa = 4.57MPASSMQMNAPSGGIAVASVNSEE138 pKa = 3.69VARR141 pKa = 11.84WVFALPRR148 pKa = 11.84EE149 pKa = 3.91LDD151 pKa = 3.71EE152 pKa = 6.05PIMEE156 pKa = 4.42TPVDD160 pKa = 3.88PSAPTEE166 pKa = 4.14AEE168 pKa = 3.86RR169 pKa = 11.84EE170 pKa = 4.08LARR173 pKa = 11.84VLNNTVAVGDD183 pKa = 3.97TASALSTIQEE193 pKa = 4.32EE194 pKa = 4.42PQHH197 pKa = 5.93EE198 pKa = 4.87VIAVPGTAVGVVHH211 pKa = 6.49QEE213 pKa = 3.69DD214 pKa = 3.99PAPVEE219 pKa = 4.47GSPSTRR225 pKa = 11.84DD226 pKa = 2.97AAMEE230 pKa = 4.11GDD232 pKa = 4.15AGHH235 pKa = 6.46SAHH238 pKa = 7.29AGDD241 pKa = 4.54DD242 pKa = 3.51TDD244 pKa = 5.57ASVEE248 pKa = 4.07MPTTVEE254 pKa = 3.57AAGPLMEE261 pKa = 5.71EE262 pKa = 4.97GPSMAHH268 pKa = 5.03LTSYY272 pKa = 8.94TVAMEE277 pKa = 3.71LRR279 pKa = 11.84SRR281 pKa = 11.84FGLRR285 pKa = 11.84PASPANIEE293 pKa = 3.74LGARR297 pKa = 11.84VARR300 pKa = 11.84EE301 pKa = 3.63ILKK304 pKa = 8.97TAGARR309 pKa = 11.84RR310 pKa = 11.84QDD312 pKa = 3.32NYY314 pKa = 11.58YY315 pKa = 10.59LAQQAVKK322 pKa = 10.34FFLTPTLLDD331 pKa = 3.47VVYY334 pKa = 8.98STPIRR339 pKa = 11.84DD340 pKa = 4.68FYY342 pKa = 11.93
MM1 pKa = 7.82DD2 pKa = 6.64HH3 pKa = 5.96ITKK6 pKa = 10.15FLRR9 pKa = 11.84MDD11 pKa = 4.23FSPKK15 pKa = 10.39DD16 pKa = 3.41GVLTRR21 pKa = 11.84RR22 pKa = 11.84QCAEE26 pKa = 4.11RR27 pKa = 11.84FDD29 pKa = 4.32DD30 pKa = 3.7TWGLLITQARR40 pKa = 11.84MTTAHH45 pKa = 6.33TADD48 pKa = 2.91MCAWYY53 pKa = 9.96EE54 pKa = 4.04GSRR57 pKa = 11.84EE58 pKa = 4.08TNQYY62 pKa = 10.81LPLEE66 pKa = 4.22QFVDD70 pKa = 3.45SRR72 pKa = 11.84DD73 pKa = 3.5ASKK76 pKa = 10.6EE77 pKa = 4.02AEE79 pKa = 4.28DD80 pKa = 4.93KK81 pKa = 11.0VDD83 pKa = 4.16KK84 pKa = 10.92LSSPPTRR91 pKa = 11.84GCGGTTVNADD101 pKa = 3.05GGLVEE106 pKa = 5.76NPLPTNRR113 pKa = 11.84DD114 pKa = 3.26EE115 pKa = 4.57MPASSMQMNAPSGGIAVASVNSEE138 pKa = 3.69VARR141 pKa = 11.84WVFALPRR148 pKa = 11.84EE149 pKa = 3.91LDD151 pKa = 3.71EE152 pKa = 6.05PIMEE156 pKa = 4.42TPVDD160 pKa = 3.88PSAPTEE166 pKa = 4.14AEE168 pKa = 3.86RR169 pKa = 11.84EE170 pKa = 4.08LARR173 pKa = 11.84VLNNTVAVGDD183 pKa = 3.97TASALSTIQEE193 pKa = 4.32EE194 pKa = 4.42PQHH197 pKa = 5.93EE198 pKa = 4.87VIAVPGTAVGVVHH211 pKa = 6.49QEE213 pKa = 3.69DD214 pKa = 3.99PAPVEE219 pKa = 4.47GSPSTRR225 pKa = 11.84DD226 pKa = 2.97AAMEE230 pKa = 4.11GDD232 pKa = 4.15AGHH235 pKa = 6.46SAHH238 pKa = 7.29AGDD241 pKa = 4.54DD242 pKa = 3.51TDD244 pKa = 5.57ASVEE248 pKa = 4.07MPTTVEE254 pKa = 3.57AAGPLMEE261 pKa = 5.71EE262 pKa = 4.97GPSMAHH268 pKa = 5.03LTSYY272 pKa = 8.94TVAMEE277 pKa = 3.71LRR279 pKa = 11.84SRR281 pKa = 11.84FGLRR285 pKa = 11.84PASPANIEE293 pKa = 3.74LGARR297 pKa = 11.84VARR300 pKa = 11.84EE301 pKa = 3.63ILKK304 pKa = 8.97TAGARR309 pKa = 11.84RR310 pKa = 11.84QDD312 pKa = 3.32NYY314 pKa = 11.58YY315 pKa = 10.59LAQQAVKK322 pKa = 10.34FFLTPTLLDD331 pKa = 3.47VVYY334 pKa = 8.98STPIRR339 pKa = 11.84DD340 pKa = 4.68FYY342 pKa = 11.93
Molecular weight: 37.02 kDa
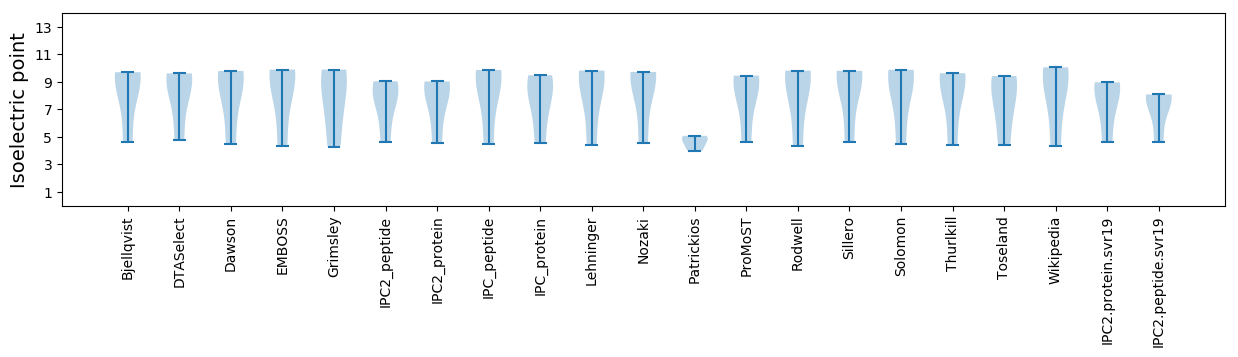
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q66129|Q66129_9TOMB Cell-to-cell movement protein OS=Carrot mottle mimic virus OX=47736 PE=4 SV=1
MM1 pKa = 7.52NLQIQPIGDD10 pKa = 3.89GNNSSRR16 pKa = 11.84NSRR19 pKa = 11.84RR20 pKa = 11.84PNQPSPRR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84DD30 pKa = 3.3PRR32 pKa = 11.84QASGSWLWCPNTSPRR47 pKa = 11.84QQYY50 pKa = 9.09PYY52 pKa = 9.45MYY54 pKa = 9.81ATIPPRR60 pKa = 11.84APTNGPGFLYY70 pKa = 10.41PEE72 pKa = 4.48MAHH75 pKa = 5.67EE76 pKa = 4.73AQWGSPVYY84 pKa = 10.4RR85 pKa = 11.84EE86 pKa = 4.1ASSGIPATRR95 pKa = 11.84ARR97 pKa = 11.84RR98 pKa = 11.84NRR100 pKa = 11.84GRR102 pKa = 11.84GSVVAPRR109 pKa = 11.84YY110 pKa = 9.95GPVGPGTGRR119 pKa = 11.84PQSVTEE125 pKa = 4.11PQRR128 pKa = 11.84GTAAGRR134 pKa = 11.84FLSEE138 pKa = 3.92LLYY141 pKa = 10.72SFEE144 pKa = 4.37RR145 pKa = 11.84FGRR148 pKa = 11.84EE149 pKa = 3.76CPAMFLPSDD158 pKa = 3.79PASRR162 pKa = 11.84HH163 pKa = 5.55PPEE166 pKa = 5.54DD167 pKa = 2.8WLQRR171 pKa = 11.84LLPALNVEE179 pKa = 4.13PSHH182 pKa = 7.02RR183 pKa = 11.84RR184 pKa = 11.84EE185 pKa = 4.31GPTLPSNRR193 pKa = 11.84AGVCAHH199 pKa = 6.67SKK201 pKa = 10.89ALDD204 pKa = 3.57KK205 pKa = 11.21EE206 pKa = 4.5LVADD210 pKa = 4.29TGTTAAVSEE219 pKa = 4.32RR220 pKa = 11.84RR221 pKa = 11.84ADD223 pKa = 3.47EE224 pKa = 4.03PTRR227 pKa = 11.84KK228 pKa = 9.58EE229 pKa = 4.17PEE231 pKa = 4.08GQPITEE237 pKa = 4.4APSTGLCSKK246 pKa = 9.62PHH248 pKa = 5.59TRR250 pKa = 11.84GEE252 pKa = 4.23QCSDD256 pKa = 3.34PTLHH260 pKa = 6.79FF261 pKa = 4.51
MM1 pKa = 7.52NLQIQPIGDD10 pKa = 3.89GNNSSRR16 pKa = 11.84NSRR19 pKa = 11.84RR20 pKa = 11.84PNQPSPRR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84DD30 pKa = 3.3PRR32 pKa = 11.84QASGSWLWCPNTSPRR47 pKa = 11.84QQYY50 pKa = 9.09PYY52 pKa = 9.45MYY54 pKa = 9.81ATIPPRR60 pKa = 11.84APTNGPGFLYY70 pKa = 10.41PEE72 pKa = 4.48MAHH75 pKa = 5.67EE76 pKa = 4.73AQWGSPVYY84 pKa = 10.4RR85 pKa = 11.84EE86 pKa = 4.1ASSGIPATRR95 pKa = 11.84ARR97 pKa = 11.84RR98 pKa = 11.84NRR100 pKa = 11.84GRR102 pKa = 11.84GSVVAPRR109 pKa = 11.84YY110 pKa = 9.95GPVGPGTGRR119 pKa = 11.84PQSVTEE125 pKa = 4.11PQRR128 pKa = 11.84GTAAGRR134 pKa = 11.84FLSEE138 pKa = 3.92LLYY141 pKa = 10.72SFEE144 pKa = 4.37RR145 pKa = 11.84FGRR148 pKa = 11.84EE149 pKa = 3.76CPAMFLPSDD158 pKa = 3.79PASRR162 pKa = 11.84HH163 pKa = 5.55PPEE166 pKa = 5.54DD167 pKa = 2.8WLQRR171 pKa = 11.84LLPALNVEE179 pKa = 4.13PSHH182 pKa = 7.02RR183 pKa = 11.84RR184 pKa = 11.84EE185 pKa = 4.31GPTLPSNRR193 pKa = 11.84AGVCAHH199 pKa = 6.67SKK201 pKa = 10.89ALDD204 pKa = 3.57KK205 pKa = 11.21EE206 pKa = 4.5LVADD210 pKa = 4.29TGTTAAVSEE219 pKa = 4.32RR220 pKa = 11.84RR221 pKa = 11.84ADD223 pKa = 3.47EE224 pKa = 4.03PTRR227 pKa = 11.84KK228 pKa = 9.58EE229 pKa = 4.17PEE231 pKa = 4.08GQPITEE237 pKa = 4.4APSTGLCSKK246 pKa = 9.62PHH248 pKa = 5.59TRR250 pKa = 11.84GEE252 pKa = 4.23QCSDD256 pKa = 3.34PTLHH260 pKa = 6.79FF261 pKa = 4.51
Molecular weight: 28.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1434 |
261 |
569 |
358.5 |
39.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.229 ± 1.275 | 1.953 ± 0.532 |
5.23 ± 0.81 | 5.718 ± 0.871 |
3.766 ± 0.81 | 7.043 ± 0.312 |
2.301 ± 0.204 | 3.696 ± 0.668 |
3.417 ± 0.908 | 8.368 ± 1.294 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.371 ± 0.563 | 3.487 ± 0.432 |
8.298 ± 1.159 | 3.975 ± 0.44 |
7.95 ± 0.791 | 7.462 ± 0.963 |
5.858 ± 1.024 | 7.043 ± 1.097 |
1.255 ± 0.125 | 2.58 ± 0.166 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
