
Propionibacterium phage E6
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Doucettevirus; Propionibacterium virus E6
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
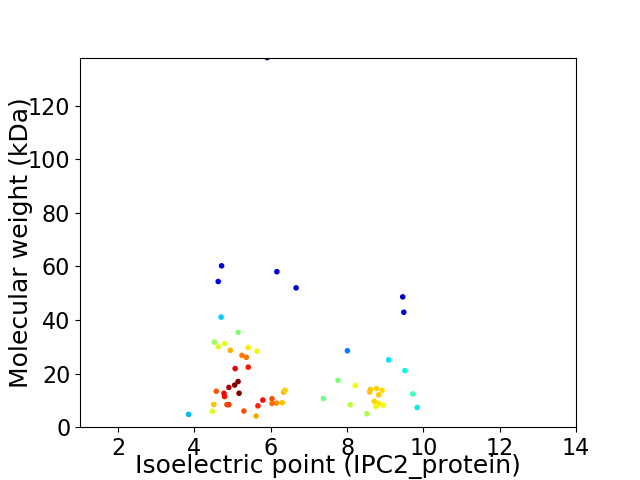
Virtual 2D-PAGE plot for 60 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1D8EU62|A0A1D8EU62_9CAUD Tape measure protein OS=Propionibacterium phage E6 OX=1897536 GN=17 PE=4 SV=1
MM1 pKa = 6.9VAWDD5 pKa = 3.8EE6 pKa = 3.97ADD8 pKa = 3.44EE9 pKa = 4.52RR10 pKa = 11.84LDD12 pKa = 3.82TGWMQARR19 pKa = 11.84CPDD22 pKa = 4.13CKK24 pKa = 10.52CFGWIAPDD32 pKa = 3.55EE33 pKa = 4.56EE34 pKa = 5.14GEE36 pKa = 4.3EE37 pKa = 4.48TNGTEE42 pKa = 4.11
MM1 pKa = 6.9VAWDD5 pKa = 3.8EE6 pKa = 3.97ADD8 pKa = 3.44EE9 pKa = 4.52RR10 pKa = 11.84LDD12 pKa = 3.82TGWMQARR19 pKa = 11.84CPDD22 pKa = 4.13CKK24 pKa = 10.52CFGWIAPDD32 pKa = 3.55EE33 pKa = 4.56EE34 pKa = 5.14GEE36 pKa = 4.3EE37 pKa = 4.48TNGTEE42 pKa = 4.11
Molecular weight: 4.79 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1D8EU25|A0A1D8EU25_9CAUD Minor tail protein OS=Propionibacterium phage E6 OX=1897536 GN=19 PE=4 SV=1
MM1 pKa = 7.08VALSRR6 pKa = 11.84ASYY9 pKa = 11.56GDD11 pKa = 3.3GTQPTQRR18 pKa = 11.84SDD20 pKa = 3.57GRR22 pKa = 11.84WAASAYY28 pKa = 10.38DD29 pKa = 3.54GWQANGNRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84WVYY43 pKa = 10.21GRR45 pKa = 11.84TQAEE49 pKa = 4.87CKK51 pKa = 10.07RR52 pKa = 11.84KK53 pKa = 9.86LRR55 pKa = 11.84DD56 pKa = 3.38LKK58 pKa = 11.11RR59 pKa = 11.84EE60 pKa = 3.81IWSDD64 pKa = 3.47TQQVNVNPRR73 pKa = 11.84EE74 pKa = 4.29TVKK77 pKa = 10.51SWTASWLDD85 pKa = 3.63DD86 pKa = 3.51YY87 pKa = 11.7RR88 pKa = 11.84SIARR92 pKa = 11.84PTTFATDD99 pKa = 2.96EE100 pKa = 4.36SMVRR104 pKa = 11.84KK105 pKa = 9.51WIVPAVGARR114 pKa = 11.84RR115 pKa = 11.84LSEE118 pKa = 3.79LTARR122 pKa = 11.84DD123 pKa = 3.26ASKK126 pKa = 10.5LQRR129 pKa = 11.84VCRR132 pKa = 11.84DD133 pKa = 3.13GGLSATTSHH142 pKa = 6.62YY143 pKa = 11.24AGLLLRR149 pKa = 11.84RR150 pKa = 11.84ILKK153 pKa = 9.78AARR156 pKa = 11.84ASGYY160 pKa = 10.03RR161 pKa = 11.84IPDD164 pKa = 3.24SVMLARR170 pKa = 11.84IPGIGASNRR179 pKa = 11.84IALSAIQAANLLSTANARR197 pKa = 11.84DD198 pKa = 3.61TWPDD202 pKa = 3.59LPSLPDD208 pKa = 3.57LPYY211 pKa = 11.22GSISKK216 pKa = 9.49LAPGEE221 pKa = 4.02AQKK224 pKa = 10.87RR225 pKa = 11.84EE226 pKa = 3.97QLKK229 pKa = 9.04MEE231 pKa = 4.34RR232 pKa = 11.84LEE234 pKa = 3.8WTAARR239 pKa = 11.84NTDD242 pKa = 3.43PSRR245 pKa = 11.84WAAALMQGLRR255 pKa = 11.84SGEE258 pKa = 3.94ARR260 pKa = 11.84GLTWDD265 pKa = 4.96RR266 pKa = 11.84VDD268 pKa = 4.48LDD270 pKa = 4.23KK271 pKa = 11.32GTITIDD277 pKa = 3.3RR278 pKa = 11.84QLQRR282 pKa = 11.84IKK284 pKa = 10.38PDD286 pKa = 3.11AALPPGYY293 pKa = 10.11KK294 pKa = 8.64VTRR297 pKa = 11.84LEE299 pKa = 4.43GSHH302 pKa = 6.67CLVAPKK308 pKa = 10.1SRR310 pKa = 11.84SGIRR314 pKa = 11.84RR315 pKa = 11.84VPIVPWMGQALTRR328 pKa = 11.84WRR330 pKa = 11.84DD331 pKa = 3.37IQGDD335 pKa = 4.21SPFGLVWPLPTGAPPTRR352 pKa = 11.84VHH354 pKa = 7.58DD355 pKa = 3.95LRR357 pKa = 11.84AWRR360 pKa = 11.84GLQRR364 pKa = 11.84VAGVHH369 pKa = 6.13KK370 pKa = 10.26EE371 pKa = 3.97DD372 pKa = 3.36GSLYY376 pKa = 8.97VLHH379 pKa = 6.94EE380 pKa = 4.58ARR382 pKa = 11.84HH383 pKa = 5.11STVSLLLAAGVPEE396 pKa = 4.29SVVIAIVGHH405 pKa = 6.51ASFAATEE412 pKa = 4.82HH413 pKa = 6.19YY414 pKa = 11.33AHH416 pKa = 7.19TDD418 pKa = 2.9LDD420 pKa = 3.85AARR423 pKa = 11.84AALMKK428 pKa = 10.61VQDD431 pKa = 3.76RR432 pKa = 11.84LGLEE436 pKa = 4.29LEE438 pKa = 4.52SS439 pKa = 4.04
MM1 pKa = 7.08VALSRR6 pKa = 11.84ASYY9 pKa = 11.56GDD11 pKa = 3.3GTQPTQRR18 pKa = 11.84SDD20 pKa = 3.57GRR22 pKa = 11.84WAASAYY28 pKa = 10.38DD29 pKa = 3.54GWQANGNRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84WVYY43 pKa = 10.21GRR45 pKa = 11.84TQAEE49 pKa = 4.87CKK51 pKa = 10.07RR52 pKa = 11.84KK53 pKa = 9.86LRR55 pKa = 11.84DD56 pKa = 3.38LKK58 pKa = 11.11RR59 pKa = 11.84EE60 pKa = 3.81IWSDD64 pKa = 3.47TQQVNVNPRR73 pKa = 11.84EE74 pKa = 4.29TVKK77 pKa = 10.51SWTASWLDD85 pKa = 3.63DD86 pKa = 3.51YY87 pKa = 11.7RR88 pKa = 11.84SIARR92 pKa = 11.84PTTFATDD99 pKa = 2.96EE100 pKa = 4.36SMVRR104 pKa = 11.84KK105 pKa = 9.51WIVPAVGARR114 pKa = 11.84RR115 pKa = 11.84LSEE118 pKa = 3.79LTARR122 pKa = 11.84DD123 pKa = 3.26ASKK126 pKa = 10.5LQRR129 pKa = 11.84VCRR132 pKa = 11.84DD133 pKa = 3.13GGLSATTSHH142 pKa = 6.62YY143 pKa = 11.24AGLLLRR149 pKa = 11.84RR150 pKa = 11.84ILKK153 pKa = 9.78AARR156 pKa = 11.84ASGYY160 pKa = 10.03RR161 pKa = 11.84IPDD164 pKa = 3.24SVMLARR170 pKa = 11.84IPGIGASNRR179 pKa = 11.84IALSAIQAANLLSTANARR197 pKa = 11.84DD198 pKa = 3.61TWPDD202 pKa = 3.59LPSLPDD208 pKa = 3.57LPYY211 pKa = 11.22GSISKK216 pKa = 9.49LAPGEE221 pKa = 4.02AQKK224 pKa = 10.87RR225 pKa = 11.84EE226 pKa = 3.97QLKK229 pKa = 9.04MEE231 pKa = 4.34RR232 pKa = 11.84LEE234 pKa = 3.8WTAARR239 pKa = 11.84NTDD242 pKa = 3.43PSRR245 pKa = 11.84WAAALMQGLRR255 pKa = 11.84SGEE258 pKa = 3.94ARR260 pKa = 11.84GLTWDD265 pKa = 4.96RR266 pKa = 11.84VDD268 pKa = 4.48LDD270 pKa = 4.23KK271 pKa = 11.32GTITIDD277 pKa = 3.3RR278 pKa = 11.84QLQRR282 pKa = 11.84IKK284 pKa = 10.38PDD286 pKa = 3.11AALPPGYY293 pKa = 10.11KK294 pKa = 8.64VTRR297 pKa = 11.84LEE299 pKa = 4.43GSHH302 pKa = 6.67CLVAPKK308 pKa = 10.1SRR310 pKa = 11.84SGIRR314 pKa = 11.84RR315 pKa = 11.84VPIVPWMGQALTRR328 pKa = 11.84WRR330 pKa = 11.84DD331 pKa = 3.37IQGDD335 pKa = 4.21SPFGLVWPLPTGAPPTRR352 pKa = 11.84VHH354 pKa = 7.58DD355 pKa = 3.95LRR357 pKa = 11.84AWRR360 pKa = 11.84GLQRR364 pKa = 11.84VAGVHH369 pKa = 6.13KK370 pKa = 10.26EE371 pKa = 3.97DD372 pKa = 3.36GSLYY376 pKa = 8.97VLHH379 pKa = 6.94EE380 pKa = 4.58ARR382 pKa = 11.84HH383 pKa = 5.11STVSLLLAAGVPEE396 pKa = 4.29SVVIAIVGHH405 pKa = 6.51ASFAATEE412 pKa = 4.82HH413 pKa = 6.19YY414 pKa = 11.33AHH416 pKa = 7.19TDD418 pKa = 2.9LDD420 pKa = 3.85AARR423 pKa = 11.84AALMKK428 pKa = 10.61VQDD431 pKa = 3.76RR432 pKa = 11.84LGLEE436 pKa = 4.29LEE438 pKa = 4.52SS439 pKa = 4.04
Molecular weight: 48.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11911 |
38 |
1364 |
198.5 |
21.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.795 ± 0.488 | 0.898 ± 0.141 |
6.238 ± 0.377 | 5.188 ± 0.449 |
2.628 ± 0.216 | 8.496 ± 0.537 |
1.813 ± 0.207 | 4.66 ± 0.403 |
3.526 ± 0.279 | 8.37 ± 0.353 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.317 ± 0.114 | 2.049 ± 0.171 |
5.608 ± 0.337 | 3.929 ± 0.213 |
7.111 ± 0.653 | 6.33 ± 0.334 |
6.716 ± 0.406 | 7.287 ± 0.381 |
2.586 ± 0.183 | 1.452 ± 0.148 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
