
Lentimonas sp. CC11
Taxonomy: cellular organisms; Bacteria; PVC group; Verrucomicrobia; Verrucomicrobia incertae sedis; Lentimonas; unclassified Lentimonas
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
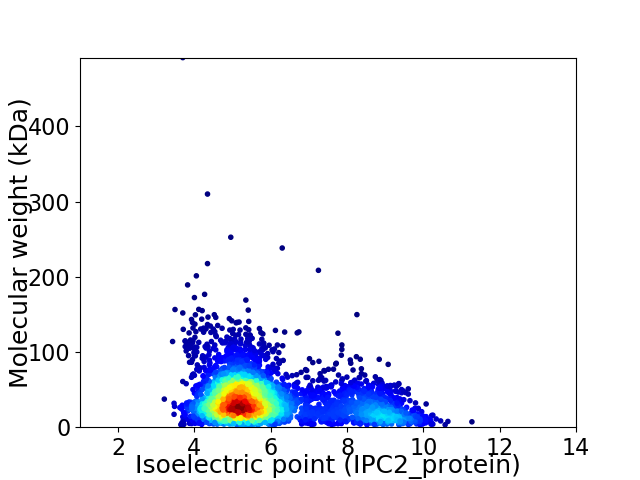
Virtual 2D-PAGE plot for 4076 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
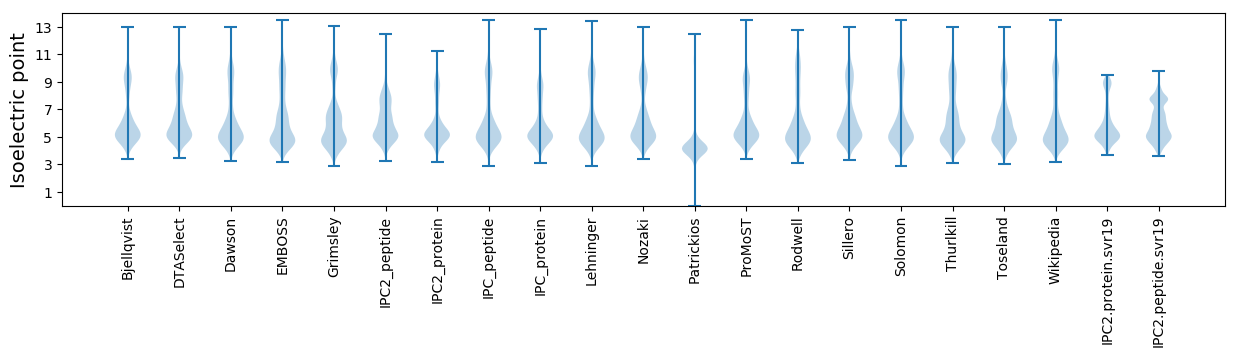
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6N4WXT5|A0A6N4WXT5_9BACT Unannotated OS=Lentimonas sp. CC11 OX=2676096 GN=LCC11_48_8 PE=4 SV=1
MM1 pKa = 6.73VCLRR5 pKa = 11.84VADD8 pKa = 4.09SATLFHH14 pKa = 6.54QSPAHH19 pKa = 4.73SVCRR23 pKa = 11.84TFVFPIFFHH32 pKa = 5.78VLPLNRR38 pKa = 11.84HH39 pKa = 6.32FIFSITFLALLSVQASDD56 pKa = 3.73TISVNFHH63 pKa = 6.39VGDD66 pKa = 4.49DD67 pKa = 3.71SDD69 pKa = 4.09NQVDD73 pKa = 4.46HH74 pKa = 7.04ILTSGEE80 pKa = 4.05MAGLASYY87 pKa = 8.37ATDD90 pKa = 2.65QWNNIDD96 pKa = 3.91IGNGGSNDD104 pKa = 3.36SAQIFAATNLSDD116 pKa = 3.65ASGNLSVATLSTSADD131 pKa = 3.74STWFTGYY138 pKa = 10.74AAGPASSEE146 pKa = 4.29AEE148 pKa = 4.0LGLAGNDD155 pKa = 2.99DD156 pKa = 4.68DD157 pKa = 6.37LFNSYY162 pKa = 10.38LALNGPSGDD171 pKa = 3.77GTPADD176 pKa = 3.54AAILNVSGLGSDD188 pKa = 3.99YY189 pKa = 11.11TSGGYY194 pKa = 10.03AVIVYY199 pKa = 10.12SDD201 pKa = 3.63SDD203 pKa = 3.75KK204 pKa = 11.01TSSSSTRR211 pKa = 11.84TSTFTLTPLGEE222 pKa = 4.15SSIALSTEE230 pKa = 3.98DD231 pKa = 3.28NATFSEE237 pKa = 4.95SYY239 pKa = 10.71DD240 pKa = 3.54ADD242 pKa = 4.19GGDD245 pKa = 3.75GYY247 pKa = 11.59SNYY250 pKa = 9.89VVFEE254 pKa = 4.11NLSAASFSLEE264 pKa = 3.94LTSPGGGGRR273 pKa = 11.84AALSGFQIIPEE284 pKa = 4.18PTISTLDD291 pKa = 3.18IVSFTTDD298 pKa = 3.18DD299 pKa = 3.64DD300 pKa = 5.32SIVVGEE306 pKa = 4.36STTLSWQTVSATSLTLNPGSVDD328 pKa = 3.37VTGTTSRR335 pKa = 11.84VVSPLATTEE344 pKa = 3.97YY345 pKa = 10.77TLIASDD351 pKa = 3.66GAGIVDD357 pKa = 4.1ASIDD361 pKa = 3.54IAVGEE366 pKa = 4.37PVPEE370 pKa = 4.42VISFTADD377 pKa = 2.78APYY380 pKa = 10.66LVVGEE385 pKa = 4.4SVVLRR390 pKa = 11.84WEE392 pKa = 3.85TAYY395 pKa = 9.7ATTLTLDD402 pKa = 3.57PGGIDD407 pKa = 3.39VTGTTSLEE415 pKa = 3.9LTPTVEE421 pKa = 4.01TTYY424 pKa = 11.08TLTASDD430 pKa = 4.07GALNDD435 pKa = 4.52DD436 pKa = 4.13ASTTIKK442 pKa = 10.49FMPAVPNILIFLVDD456 pKa = 4.52DD457 pKa = 3.82MGITDD462 pKa = 3.74TSVPFVYY469 pKa = 10.47DD470 pKa = 3.26EE471 pKa = 5.55GVAQYY476 pKa = 11.82YY477 pKa = 9.49NFNYY481 pKa = 10.1YY482 pKa = 10.19NHH484 pKa = 6.67TPNMEE489 pKa = 4.22TLASQGMKK497 pKa = 9.22FTQAYY502 pKa = 6.05ATPVCSSTRR511 pKa = 11.84VSLMSGYY518 pKa = 8.24NTTRR522 pKa = 11.84HH523 pKa = 5.14GVISQVPPSGTEE535 pKa = 3.47PATISPVTSLLDD547 pKa = 3.81PPNNWKK553 pKa = 8.78RR554 pKa = 11.84TGLLEE559 pKa = 4.18SDD561 pKa = 4.82KK562 pKa = 10.91EE563 pKa = 5.11SMMPWILRR571 pKa = 11.84EE572 pKa = 3.66AGYY575 pKa = 8.23RR576 pKa = 11.84TIHH579 pKa = 6.43AGKK582 pKa = 10.01AHH584 pKa = 6.71FSSTNGFAKK593 pKa = 10.56YY594 pKa = 7.42PTAIGFDD601 pKa = 3.76VNLGGTQAGSSNYY614 pKa = 8.65TNYY617 pKa = 10.88GGLPNMSSYY626 pKa = 11.22AAADD630 pKa = 3.57VFLTDD635 pKa = 5.42ALTQAMNSEE644 pKa = 4.4IGNSLEE650 pKa = 4.01AGLPFFAYY658 pKa = 7.98MSHH661 pKa = 6.8YY662 pKa = 9.91GVHH665 pKa = 6.37SPFTNDD671 pKa = 2.81PNATADD677 pKa = 3.42YY678 pKa = 9.35TKK680 pKa = 10.38IVLPDD685 pKa = 3.43EE686 pKa = 4.97SEE688 pKa = 4.41VSVSNSNQRR697 pKa = 11.84IFGSMLEE704 pKa = 4.68GMDD707 pKa = 3.48QSLGSILEE715 pKa = 3.93QLDD718 pKa = 3.56ALGVAEE724 pKa = 4.22EE725 pKa = 4.25TLVIFMGDD733 pKa = 3.43NGSDD737 pKa = 3.56APQATYY743 pKa = 11.07NGFAGSPFSDD753 pKa = 3.94FPLRR757 pKa = 11.84GKK759 pKa = 10.28KK760 pKa = 9.95GSSWEE765 pKa = 3.88GGIRR769 pKa = 11.84VPMIVAWAKK778 pKa = 10.28VDD780 pKa = 3.42PTNPVQAAFPIPQGTVEE797 pKa = 4.74DD798 pKa = 4.44DD799 pKa = 4.57LVVVWDD805 pKa = 4.14ILPTVLNITEE815 pKa = 4.09VEE817 pKa = 4.17APHH820 pKa = 7.24VFDD823 pKa = 6.41GYY825 pKa = 11.5DD826 pKa = 3.34LTPYY830 pKa = 10.97LLGHH834 pKa = 7.29AGTHH838 pKa = 6.38RR839 pKa = 11.84PQEE842 pKa = 3.98MLMYY846 pKa = 10.65LPIDD850 pKa = 3.38HH851 pKa = 7.09RR852 pKa = 11.84NDD854 pKa = 3.03YY855 pKa = 10.73FSIYY859 pKa = 9.84RR860 pKa = 11.84DD861 pKa = 3.87EE862 pKa = 4.16EE863 pKa = 3.8WKK865 pKa = 10.76LIYY868 pKa = 10.46FFATDD873 pKa = 4.14SYY875 pKa = 11.46TLYY878 pKa = 11.29NLDD881 pKa = 3.65TDD883 pKa = 3.98PTEE886 pKa = 4.9SNDD889 pKa = 3.34VAGDD893 pKa = 3.34HH894 pKa = 6.84SDD896 pKa = 3.17RR897 pKa = 11.84VMAMARR903 pKa = 11.84SMAQEE908 pKa = 4.47FEE910 pKa = 4.53RR911 pKa = 11.84TWGDD915 pKa = 3.96QGQLWPTFDD924 pKa = 4.69SDD926 pKa = 4.7SSDD929 pKa = 4.88DD930 pKa = 4.0PFSMPNLPAVDD941 pKa = 4.94LDD943 pKa = 3.71MDD945 pKa = 4.25GLGDD949 pKa = 3.69NSEE952 pKa = 4.26DD953 pKa = 3.6ANLNGLIDD961 pKa = 4.56AGEE964 pKa = 4.27TDD966 pKa = 4.08PDD968 pKa = 3.71DD969 pKa = 3.59WDD971 pKa = 3.82TDD973 pKa = 3.49GDD975 pKa = 3.86RR976 pKa = 11.84TDD978 pKa = 5.35DD979 pKa = 3.57YY980 pKa = 11.46TEE982 pKa = 4.01RR983 pKa = 11.84RR984 pKa = 11.84LNLDD988 pKa = 3.67PLDD991 pKa = 4.3ANSFFAARR999 pKa = 11.84VVSTGTGNLDD1009 pKa = 3.9LAWPSAPGLSFDD1021 pKa = 4.82ILSSDD1026 pKa = 4.13DD1027 pKa = 3.78LSTPFEE1033 pKa = 4.32DD1034 pKa = 3.06WTRR1037 pKa = 11.84LISGVEE1043 pKa = 3.84ADD1045 pKa = 4.15DD1046 pKa = 4.02TLAEE1050 pKa = 4.68TIQSLTIDD1058 pKa = 3.8PEE1060 pKa = 3.99QDD1062 pKa = 2.87KK1063 pKa = 10.91EE1064 pKa = 4.48FYY1066 pKa = 10.61SVEE1069 pKa = 4.02LLPP1072 pKa = 5.54
MM1 pKa = 6.73VCLRR5 pKa = 11.84VADD8 pKa = 4.09SATLFHH14 pKa = 6.54QSPAHH19 pKa = 4.73SVCRR23 pKa = 11.84TFVFPIFFHH32 pKa = 5.78VLPLNRR38 pKa = 11.84HH39 pKa = 6.32FIFSITFLALLSVQASDD56 pKa = 3.73TISVNFHH63 pKa = 6.39VGDD66 pKa = 4.49DD67 pKa = 3.71SDD69 pKa = 4.09NQVDD73 pKa = 4.46HH74 pKa = 7.04ILTSGEE80 pKa = 4.05MAGLASYY87 pKa = 8.37ATDD90 pKa = 2.65QWNNIDD96 pKa = 3.91IGNGGSNDD104 pKa = 3.36SAQIFAATNLSDD116 pKa = 3.65ASGNLSVATLSTSADD131 pKa = 3.74STWFTGYY138 pKa = 10.74AAGPASSEE146 pKa = 4.29AEE148 pKa = 4.0LGLAGNDD155 pKa = 2.99DD156 pKa = 4.68DD157 pKa = 6.37LFNSYY162 pKa = 10.38LALNGPSGDD171 pKa = 3.77GTPADD176 pKa = 3.54AAILNVSGLGSDD188 pKa = 3.99YY189 pKa = 11.11TSGGYY194 pKa = 10.03AVIVYY199 pKa = 10.12SDD201 pKa = 3.63SDD203 pKa = 3.75KK204 pKa = 11.01TSSSSTRR211 pKa = 11.84TSTFTLTPLGEE222 pKa = 4.15SSIALSTEE230 pKa = 3.98DD231 pKa = 3.28NATFSEE237 pKa = 4.95SYY239 pKa = 10.71DD240 pKa = 3.54ADD242 pKa = 4.19GGDD245 pKa = 3.75GYY247 pKa = 11.59SNYY250 pKa = 9.89VVFEE254 pKa = 4.11NLSAASFSLEE264 pKa = 3.94LTSPGGGGRR273 pKa = 11.84AALSGFQIIPEE284 pKa = 4.18PTISTLDD291 pKa = 3.18IVSFTTDD298 pKa = 3.18DD299 pKa = 3.64DD300 pKa = 5.32SIVVGEE306 pKa = 4.36STTLSWQTVSATSLTLNPGSVDD328 pKa = 3.37VTGTTSRR335 pKa = 11.84VVSPLATTEE344 pKa = 3.97YY345 pKa = 10.77TLIASDD351 pKa = 3.66GAGIVDD357 pKa = 4.1ASIDD361 pKa = 3.54IAVGEE366 pKa = 4.37PVPEE370 pKa = 4.42VISFTADD377 pKa = 2.78APYY380 pKa = 10.66LVVGEE385 pKa = 4.4SVVLRR390 pKa = 11.84WEE392 pKa = 3.85TAYY395 pKa = 9.7ATTLTLDD402 pKa = 3.57PGGIDD407 pKa = 3.39VTGTTSLEE415 pKa = 3.9LTPTVEE421 pKa = 4.01TTYY424 pKa = 11.08TLTASDD430 pKa = 4.07GALNDD435 pKa = 4.52DD436 pKa = 4.13ASTTIKK442 pKa = 10.49FMPAVPNILIFLVDD456 pKa = 4.52DD457 pKa = 3.82MGITDD462 pKa = 3.74TSVPFVYY469 pKa = 10.47DD470 pKa = 3.26EE471 pKa = 5.55GVAQYY476 pKa = 11.82YY477 pKa = 9.49NFNYY481 pKa = 10.1YY482 pKa = 10.19NHH484 pKa = 6.67TPNMEE489 pKa = 4.22TLASQGMKK497 pKa = 9.22FTQAYY502 pKa = 6.05ATPVCSSTRR511 pKa = 11.84VSLMSGYY518 pKa = 8.24NTTRR522 pKa = 11.84HH523 pKa = 5.14GVISQVPPSGTEE535 pKa = 3.47PATISPVTSLLDD547 pKa = 3.81PPNNWKK553 pKa = 8.78RR554 pKa = 11.84TGLLEE559 pKa = 4.18SDD561 pKa = 4.82KK562 pKa = 10.91EE563 pKa = 5.11SMMPWILRR571 pKa = 11.84EE572 pKa = 3.66AGYY575 pKa = 8.23RR576 pKa = 11.84TIHH579 pKa = 6.43AGKK582 pKa = 10.01AHH584 pKa = 6.71FSSTNGFAKK593 pKa = 10.56YY594 pKa = 7.42PTAIGFDD601 pKa = 3.76VNLGGTQAGSSNYY614 pKa = 8.65TNYY617 pKa = 10.88GGLPNMSSYY626 pKa = 11.22AAADD630 pKa = 3.57VFLTDD635 pKa = 5.42ALTQAMNSEE644 pKa = 4.4IGNSLEE650 pKa = 4.01AGLPFFAYY658 pKa = 7.98MSHH661 pKa = 6.8YY662 pKa = 9.91GVHH665 pKa = 6.37SPFTNDD671 pKa = 2.81PNATADD677 pKa = 3.42YY678 pKa = 9.35TKK680 pKa = 10.38IVLPDD685 pKa = 3.43EE686 pKa = 4.97SEE688 pKa = 4.41VSVSNSNQRR697 pKa = 11.84IFGSMLEE704 pKa = 4.68GMDD707 pKa = 3.48QSLGSILEE715 pKa = 3.93QLDD718 pKa = 3.56ALGVAEE724 pKa = 4.22EE725 pKa = 4.25TLVIFMGDD733 pKa = 3.43NGSDD737 pKa = 3.56APQATYY743 pKa = 11.07NGFAGSPFSDD753 pKa = 3.94FPLRR757 pKa = 11.84GKK759 pKa = 10.28KK760 pKa = 9.95GSSWEE765 pKa = 3.88GGIRR769 pKa = 11.84VPMIVAWAKK778 pKa = 10.28VDD780 pKa = 3.42PTNPVQAAFPIPQGTVEE797 pKa = 4.74DD798 pKa = 4.44DD799 pKa = 4.57LVVVWDD805 pKa = 4.14ILPTVLNITEE815 pKa = 4.09VEE817 pKa = 4.17APHH820 pKa = 7.24VFDD823 pKa = 6.41GYY825 pKa = 11.5DD826 pKa = 3.34LTPYY830 pKa = 10.97LLGHH834 pKa = 7.29AGTHH838 pKa = 6.38RR839 pKa = 11.84PQEE842 pKa = 3.98MLMYY846 pKa = 10.65LPIDD850 pKa = 3.38HH851 pKa = 7.09RR852 pKa = 11.84NDD854 pKa = 3.03YY855 pKa = 10.73FSIYY859 pKa = 9.84RR860 pKa = 11.84DD861 pKa = 3.87EE862 pKa = 4.16EE863 pKa = 3.8WKK865 pKa = 10.76LIYY868 pKa = 10.46FFATDD873 pKa = 4.14SYY875 pKa = 11.46TLYY878 pKa = 11.29NLDD881 pKa = 3.65TDD883 pKa = 3.98PTEE886 pKa = 4.9SNDD889 pKa = 3.34VAGDD893 pKa = 3.34HH894 pKa = 6.84SDD896 pKa = 3.17RR897 pKa = 11.84VMAMARR903 pKa = 11.84SMAQEE908 pKa = 4.47FEE910 pKa = 4.53RR911 pKa = 11.84TWGDD915 pKa = 3.96QGQLWPTFDD924 pKa = 4.69SDD926 pKa = 4.7SSDD929 pKa = 4.88DD930 pKa = 4.0PFSMPNLPAVDD941 pKa = 4.94LDD943 pKa = 3.71MDD945 pKa = 4.25GLGDD949 pKa = 3.69NSEE952 pKa = 4.26DD953 pKa = 3.6ANLNGLIDD961 pKa = 4.56AGEE964 pKa = 4.27TDD966 pKa = 4.08PDD968 pKa = 3.71DD969 pKa = 3.59WDD971 pKa = 3.82TDD973 pKa = 3.49GDD975 pKa = 3.86RR976 pKa = 11.84TDD978 pKa = 5.35DD979 pKa = 3.57YY980 pKa = 11.46TEE982 pKa = 4.01RR983 pKa = 11.84RR984 pKa = 11.84LNLDD988 pKa = 3.67PLDD991 pKa = 4.3ANSFFAARR999 pKa = 11.84VVSTGTGNLDD1009 pKa = 3.9LAWPSAPGLSFDD1021 pKa = 4.82ILSSDD1026 pKa = 4.13DD1027 pKa = 3.78LSTPFEE1033 pKa = 4.32DD1034 pKa = 3.06WTRR1037 pKa = 11.84LISGVEE1043 pKa = 3.84ADD1045 pKa = 4.15DD1046 pKa = 4.02TLAEE1050 pKa = 4.68TIQSLTIDD1058 pKa = 3.8PEE1060 pKa = 3.99QDD1062 pKa = 2.87KK1063 pKa = 10.91EE1064 pKa = 4.48FYY1066 pKa = 10.61SVEE1069 pKa = 4.02LLPP1072 pKa = 5.54
Molecular weight: 114.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6N4WXM3|A0A6N4WXM3_9BACT Unannotated OS=Lentimonas sp. CC11 OX=2676096 GN=LCC11_26_3 PE=4 SV=1
MM1 pKa = 7.14QKK3 pKa = 8.56TRR5 pKa = 11.84KK6 pKa = 9.97SIAKK10 pKa = 9.38RR11 pKa = 11.84FKK13 pKa = 9.88KK14 pKa = 9.24TGTGKK19 pKa = 10.57LLRR22 pKa = 11.84RR23 pKa = 11.84TPGHH27 pKa = 5.98RR28 pKa = 11.84HH29 pKa = 5.44LLRR32 pKa = 11.84NKK34 pKa = 9.4SVKK37 pKa = 7.93QRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84AGSSKK46 pKa = 10.36LVAPGQRR53 pKa = 11.84ANLIRR58 pKa = 11.84GLPFAA63 pKa = 5.56
MM1 pKa = 7.14QKK3 pKa = 8.56TRR5 pKa = 11.84KK6 pKa = 9.97SIAKK10 pKa = 9.38RR11 pKa = 11.84FKK13 pKa = 9.88KK14 pKa = 9.24TGTGKK19 pKa = 10.57LLRR22 pKa = 11.84RR23 pKa = 11.84TPGHH27 pKa = 5.98RR28 pKa = 11.84HH29 pKa = 5.44LLRR32 pKa = 11.84NKK34 pKa = 9.4SVKK37 pKa = 7.93QRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84AGSSKK46 pKa = 10.36LVAPGQRR53 pKa = 11.84ANLIRR58 pKa = 11.84GLPFAA63 pKa = 5.56
Molecular weight: 7.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1428820 |
25 |
4680 |
350.5 |
38.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.328 ± 0.036 | 1.079 ± 0.014 |
5.984 ± 0.036 | 6.443 ± 0.036 |
4.137 ± 0.02 | 7.508 ± 0.04 |
2.188 ± 0.019 | 5.842 ± 0.027 |
4.667 ± 0.037 | 9.768 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.289 ± 0.019 | 3.766 ± 0.026 |
4.542 ± 0.024 | 3.688 ± 0.024 |
5.199 ± 0.039 | 6.591 ± 0.033 |
5.744 ± 0.041 | 6.671 ± 0.036 |
1.47 ± 0.016 | 3.091 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
