
Paraburkholderia susongensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Burkholderiaceae; Paraburkholderia
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
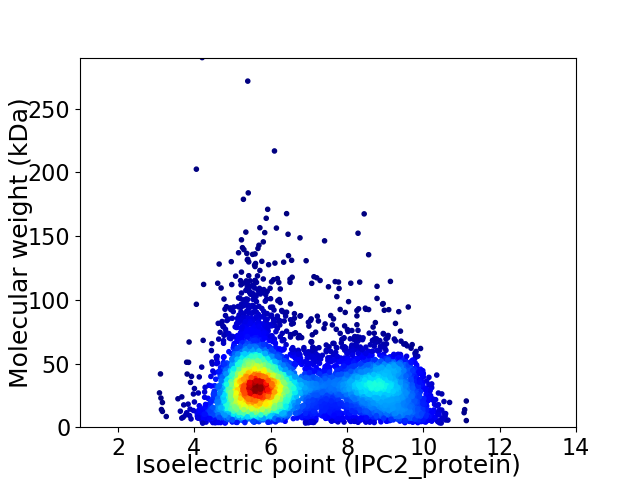
Virtual 2D-PAGE plot for 6846 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X7KCU6|A0A1X7KCU6_9BURK Methyl-accepting chemotaxis sensory transducer with TarH sensor OS=Paraburkholderia susongensis OX=1515439 GN=SAMN06265784_103569 PE=4 SV=1
MM1 pKa = 7.88RR2 pKa = 11.84ISSAQYY8 pKa = 9.39FSMNVEE14 pKa = 4.21TMSNQQAQLSQIYY27 pKa = 8.43QQVSSGQSLTTPADD41 pKa = 3.65NPLGAAQAVQLSSTAAALSQYY62 pKa = 7.34TTNQSAALASLQAEE76 pKa = 4.52DD77 pKa = 3.85STLSGVLSTLQSVNTLGIRR96 pKa = 11.84ASNGSLNDD104 pKa = 3.67ADD106 pKa = 4.0RR107 pKa = 11.84TAIAAQLTSLRR118 pKa = 11.84DD119 pKa = 3.51QLLTYY124 pKa = 11.26ANATDD129 pKa = 3.6SAGNSLFSGFQNTAQAFTTDD149 pKa = 2.66ASGAVVYY156 pKa = 10.95SGDD159 pKa = 3.31NGTRR163 pKa = 11.84TVQVTGSNQVATGDD177 pKa = 3.28NGRR180 pKa = 11.84AVFMSVGSTNAEE192 pKa = 4.1PIAAGNSANTGTGVIGAVTVNDD214 pKa = 3.69PTQVANADD222 pKa = 3.69SYY224 pKa = 11.53QISFSGSGSTLAYY237 pKa = 9.12TITDD241 pKa = 3.59TTTGVAGSPQAYY253 pKa = 8.32TEE255 pKa = 4.24GSNITLGGQTVAITGTPADD274 pKa = 3.98GDD276 pKa = 3.88NFTVKK281 pKa = 9.97PANQAGTDD289 pKa = 3.41VFANLNAMISALQQPVTGNTAAIANLQNSLNTGLSQLANTLGNVTAVQASVGGRR343 pKa = 11.84EE344 pKa = 3.98QEE346 pKa = 4.23VQALQTVTQTNSLQTQSNLSDD367 pKa = 3.47LTSTDD372 pKa = 3.23MVSAISKK379 pKa = 8.02YY380 pKa = 8.04TLQQAALQASQQAFVQIQNMSLFQYY405 pKa = 10.64INNN408 pKa = 3.99
MM1 pKa = 7.88RR2 pKa = 11.84ISSAQYY8 pKa = 9.39FSMNVEE14 pKa = 4.21TMSNQQAQLSQIYY27 pKa = 8.43QQVSSGQSLTTPADD41 pKa = 3.65NPLGAAQAVQLSSTAAALSQYY62 pKa = 7.34TTNQSAALASLQAEE76 pKa = 4.52DD77 pKa = 3.85STLSGVLSTLQSVNTLGIRR96 pKa = 11.84ASNGSLNDD104 pKa = 3.67ADD106 pKa = 4.0RR107 pKa = 11.84TAIAAQLTSLRR118 pKa = 11.84DD119 pKa = 3.51QLLTYY124 pKa = 11.26ANATDD129 pKa = 3.6SAGNSLFSGFQNTAQAFTTDD149 pKa = 2.66ASGAVVYY156 pKa = 10.95SGDD159 pKa = 3.31NGTRR163 pKa = 11.84TVQVTGSNQVATGDD177 pKa = 3.28NGRR180 pKa = 11.84AVFMSVGSTNAEE192 pKa = 4.1PIAAGNSANTGTGVIGAVTVNDD214 pKa = 3.69PTQVANADD222 pKa = 3.69SYY224 pKa = 11.53QISFSGSGSTLAYY237 pKa = 9.12TITDD241 pKa = 3.59TTTGVAGSPQAYY253 pKa = 8.32TEE255 pKa = 4.24GSNITLGGQTVAITGTPADD274 pKa = 3.98GDD276 pKa = 3.88NFTVKK281 pKa = 9.97PANQAGTDD289 pKa = 3.41VFANLNAMISALQQPVTGNTAAIANLQNSLNTGLSQLANTLGNVTAVQASVGGRR343 pKa = 11.84EE344 pKa = 3.98QEE346 pKa = 4.23VQALQTVTQTNSLQTQSNLSDD367 pKa = 3.47LTSTDD372 pKa = 3.23MVSAISKK379 pKa = 8.02YY380 pKa = 8.04TLQQAALQASQQAFVQIQNMSLFQYY405 pKa = 10.64INNN408 pKa = 3.99
Molecular weight: 41.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X7K591|A0A1X7K591_9BURK Acetylglutamate kinase OS=Paraburkholderia susongensis OX=1515439 GN=argB PE=3 SV=1
MM1 pKa = 7.11HH2 pKa = 7.28TLLSTRR8 pKa = 11.84LTRR11 pKa = 11.84ASLKK15 pKa = 9.97AARR18 pKa = 11.84PARR21 pKa = 11.84RR22 pKa = 11.84HH23 pKa = 5.14LMRR26 pKa = 11.84ALRR29 pKa = 11.84HH30 pKa = 4.9HH31 pKa = 7.27AARR34 pKa = 11.84TRR36 pKa = 11.84ALGKK40 pKa = 7.98QWRR43 pKa = 11.84DD44 pKa = 2.83ARR46 pKa = 11.84AVDD49 pKa = 5.58GIRR52 pKa = 11.84TWFHH56 pKa = 6.51AFFSALRR63 pKa = 11.84QQSGARR69 pKa = 11.84RR70 pKa = 11.84FSLRR74 pKa = 11.84TLLRR78 pKa = 11.84KK79 pKa = 7.51PTPLRR84 pKa = 11.84LVVAKK89 pKa = 9.89RR90 pKa = 11.84APATPVNTSRR100 pKa = 11.84RR101 pKa = 11.84PRR103 pKa = 11.84RR104 pKa = 11.84LSTSSSGWFAFAFASRR120 pKa = 3.71
MM1 pKa = 7.11HH2 pKa = 7.28TLLSTRR8 pKa = 11.84LTRR11 pKa = 11.84ASLKK15 pKa = 9.97AARR18 pKa = 11.84PARR21 pKa = 11.84RR22 pKa = 11.84HH23 pKa = 5.14LMRR26 pKa = 11.84ALRR29 pKa = 11.84HH30 pKa = 4.9HH31 pKa = 7.27AARR34 pKa = 11.84TRR36 pKa = 11.84ALGKK40 pKa = 7.98QWRR43 pKa = 11.84DD44 pKa = 2.83ARR46 pKa = 11.84AVDD49 pKa = 5.58GIRR52 pKa = 11.84TWFHH56 pKa = 6.51AFFSALRR63 pKa = 11.84QQSGARR69 pKa = 11.84RR70 pKa = 11.84FSLRR74 pKa = 11.84TLLRR78 pKa = 11.84KK79 pKa = 7.51PTPLRR84 pKa = 11.84LVVAKK89 pKa = 9.89RR90 pKa = 11.84APATPVNTSRR100 pKa = 11.84RR101 pKa = 11.84PRR103 pKa = 11.84RR104 pKa = 11.84LSTSSSGWFAFAFASRR120 pKa = 3.71
Molecular weight: 13.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2196354 |
29 |
2911 |
320.8 |
34.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.661 ± 0.043 | 0.951 ± 0.011 |
5.29 ± 0.026 | 5.127 ± 0.033 |
3.749 ± 0.019 | 8.194 ± 0.03 |
2.318 ± 0.015 | 4.793 ± 0.022 |
3.015 ± 0.026 | 10.251 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.413 ± 0.013 | 2.889 ± 0.022 |
5.088 ± 0.026 | 3.612 ± 0.018 |
6.881 ± 0.03 | 5.795 ± 0.025 |
5.447 ± 0.024 | 7.709 ± 0.023 |
1.37 ± 0.014 | 2.448 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
