
Chaetothyriales sp. CBS 132003
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Chaetothyriomycetidae; Chaetothyriales; unclassified Chaetothyriales
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
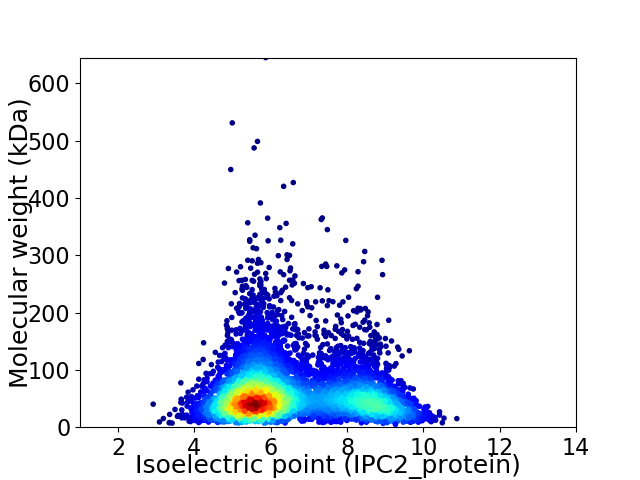
Virtual 2D-PAGE plot for 5726 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3M7MVX4|A0A3M7MVX4_9EURO WD_REPEATS_REGION domain-containing protein (Fragment) OS=Chaetothyriales sp. CBS 132003 OX=2249419 GN=DV737_g4098 PE=4 SV=1
MM1 pKa = 7.51LFSSYY6 pKa = 10.7AVQASVLLLISTVFGQYY23 pKa = 10.44SLQDD27 pKa = 3.87DD28 pKa = 4.57YY29 pKa = 10.14TTSSFGSMFDD39 pKa = 4.33FFTDD43 pKa = 3.8DD44 pKa = 4.91DD45 pKa = 4.15PTSGYY50 pKa = 10.55VNYY53 pKa = 10.01VSQSEE58 pKa = 4.53AEE60 pKa = 4.28SQGLFTTEE68 pKa = 3.7NNTVYY73 pKa = 10.43IGVDD77 pKa = 3.42STNVASGRR85 pKa = 11.84GRR87 pKa = 11.84NSVRR91 pKa = 11.84ISSKK95 pKa = 10.3NVYY98 pKa = 7.72THH100 pKa = 6.5GLIILDD106 pKa = 4.2LEE108 pKa = 4.47HH109 pKa = 6.91MPGGQCGSWPAFWTLGPDD127 pKa = 2.86WPTWGEE133 pKa = 3.1IDD135 pKa = 4.36IIEE138 pKa = 4.51NVNSATVNTISAHH151 pKa = 5.53TDD153 pKa = 3.16SSCSISATGNFTGSILTTDD172 pKa = 4.54CDD174 pKa = 3.56SSAASEE180 pKa = 4.42SSDD183 pKa = 3.51SGCSFTDD190 pKa = 3.11TSTQTFGAGFNAIGGGVYY208 pKa = 8.89ATEE211 pKa = 4.26WTSEE215 pKa = 4.31AISIWFFPRR224 pKa = 11.84LLIPSDD230 pKa = 3.41ITNGSPDD237 pKa = 3.39PSGWGEE243 pKa = 3.87PLAQFTGGGCDD254 pKa = 3.79IDD256 pKa = 5.01DD257 pKa = 3.9HH258 pKa = 6.65FKK260 pKa = 11.11NNQVVFDD267 pKa = 4.64ISMCGDD273 pKa = 2.98WAGDD277 pKa = 3.41VWSQDD282 pKa = 3.28ATCSLLASTCEE293 pKa = 5.14DD294 pKa = 3.69YY295 pKa = 11.48VQNNPDD301 pKa = 4.0AFVDD305 pKa = 4.35TYY307 pKa = 11.35WLINSLKK314 pKa = 10.38VYY316 pKa = 10.28QEE318 pKa = 4.2NNQQDD323 pKa = 4.22TTSSLSPSSTAGTTTSTASSAATITTTSSSVVPTTFEE360 pKa = 3.86TSTRR364 pKa = 11.84PIVVPSSVTSTDD376 pKa = 2.5TWSASVSIPTSTDD389 pKa = 1.95IWSASVSIPTSTDD402 pKa = 2.83APSVLEE408 pKa = 4.25STSTTTDD415 pKa = 2.98TSSVSVAGSPFATQGWQDD433 pKa = 3.7LDD435 pKa = 3.68GGKK438 pKa = 9.83QGWGLQGSGWGQRR451 pKa = 11.84NRR453 pKa = 11.84GRR455 pKa = 11.84WSGGDD460 pKa = 3.1
MM1 pKa = 7.51LFSSYY6 pKa = 10.7AVQASVLLLISTVFGQYY23 pKa = 10.44SLQDD27 pKa = 3.87DD28 pKa = 4.57YY29 pKa = 10.14TTSSFGSMFDD39 pKa = 4.33FFTDD43 pKa = 3.8DD44 pKa = 4.91DD45 pKa = 4.15PTSGYY50 pKa = 10.55VNYY53 pKa = 10.01VSQSEE58 pKa = 4.53AEE60 pKa = 4.28SQGLFTTEE68 pKa = 3.7NNTVYY73 pKa = 10.43IGVDD77 pKa = 3.42STNVASGRR85 pKa = 11.84GRR87 pKa = 11.84NSVRR91 pKa = 11.84ISSKK95 pKa = 10.3NVYY98 pKa = 7.72THH100 pKa = 6.5GLIILDD106 pKa = 4.2LEE108 pKa = 4.47HH109 pKa = 6.91MPGGQCGSWPAFWTLGPDD127 pKa = 2.86WPTWGEE133 pKa = 3.1IDD135 pKa = 4.36IIEE138 pKa = 4.51NVNSATVNTISAHH151 pKa = 5.53TDD153 pKa = 3.16SSCSISATGNFTGSILTTDD172 pKa = 4.54CDD174 pKa = 3.56SSAASEE180 pKa = 4.42SSDD183 pKa = 3.51SGCSFTDD190 pKa = 3.11TSTQTFGAGFNAIGGGVYY208 pKa = 8.89ATEE211 pKa = 4.26WTSEE215 pKa = 4.31AISIWFFPRR224 pKa = 11.84LLIPSDD230 pKa = 3.41ITNGSPDD237 pKa = 3.39PSGWGEE243 pKa = 3.87PLAQFTGGGCDD254 pKa = 3.79IDD256 pKa = 5.01DD257 pKa = 3.9HH258 pKa = 6.65FKK260 pKa = 11.11NNQVVFDD267 pKa = 4.64ISMCGDD273 pKa = 2.98WAGDD277 pKa = 3.41VWSQDD282 pKa = 3.28ATCSLLASTCEE293 pKa = 5.14DD294 pKa = 3.69YY295 pKa = 11.48VQNNPDD301 pKa = 4.0AFVDD305 pKa = 4.35TYY307 pKa = 11.35WLINSLKK314 pKa = 10.38VYY316 pKa = 10.28QEE318 pKa = 4.2NNQQDD323 pKa = 4.22TTSSLSPSSTAGTTTSTASSAATITTTSSSVVPTTFEE360 pKa = 3.86TSTRR364 pKa = 11.84PIVVPSSVTSTDD376 pKa = 2.5TWSASVSIPTSTDD389 pKa = 1.95IWSASVSIPTSTDD402 pKa = 2.83APSVLEE408 pKa = 4.25STSTTTDD415 pKa = 2.98TSSVSVAGSPFATQGWQDD433 pKa = 3.7LDD435 pKa = 3.68GGKK438 pKa = 9.83QGWGLQGSGWGQRR451 pKa = 11.84NRR453 pKa = 11.84GRR455 pKa = 11.84WSGGDD460 pKa = 3.1
Molecular weight: 48.66 kDa
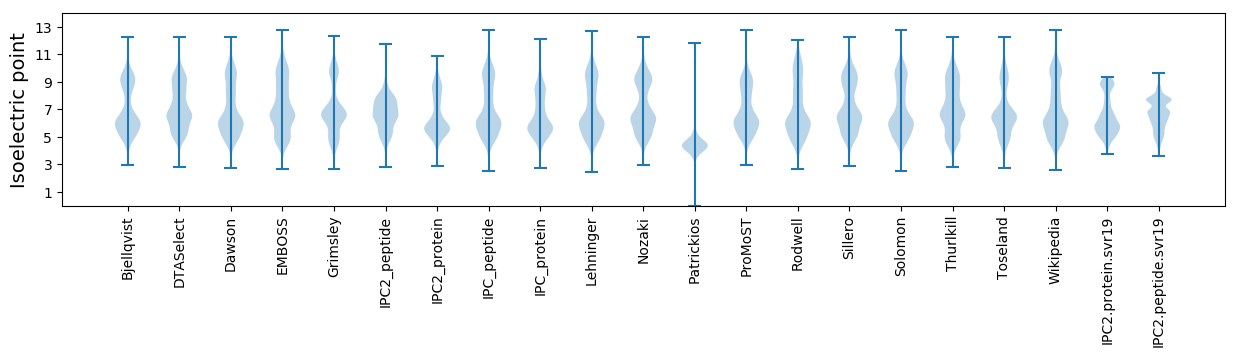
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3M7MQD0|A0A3M7MQD0_9EURO PCI domain-containing protein (Fragment) OS=Chaetothyriales sp. CBS 132003 OX=2249419 GN=DV737_g4633 PE=4 SV=1
MM1 pKa = 7.96PSTADD6 pKa = 3.45GDD8 pKa = 4.32CSQTHH13 pKa = 6.0AQTHH17 pKa = 5.2AQTRR21 pKa = 11.84VQTRR25 pKa = 11.84TSAASPNDD33 pKa = 4.06SIRR36 pKa = 11.84PPAPARR42 pKa = 11.84CSRR45 pKa = 11.84MVSRR49 pKa = 11.84CGAAEE54 pKa = 3.88RR55 pKa = 11.84TRR57 pKa = 11.84LQDD60 pKa = 3.26VVQQRR65 pKa = 11.84IARR68 pKa = 11.84LPHH71 pKa = 5.52AQQLARR77 pKa = 11.84LHH79 pKa = 6.44PKK81 pKa = 9.44RR82 pKa = 11.84AAARR86 pKa = 11.84TSSANASQPRR96 pKa = 11.84TQTPAEE102 pKa = 4.38VKK104 pKa = 9.76SQRR107 pKa = 11.84PEE109 pKa = 3.34ARR111 pKa = 11.84QTGRR115 pKa = 11.84SPRR118 pKa = 11.84EE119 pKa = 3.39EE120 pKa = 3.7WQKK123 pKa = 10.76FIHH126 pKa = 6.54RR127 pKa = 11.84LSALPRR133 pKa = 11.84TRR135 pKa = 11.84PSRR138 pKa = 11.84EE139 pKa = 3.83SNGRR143 pKa = 11.84CVEE146 pKa = 3.92LTTAFVHH153 pKa = 6.76HH154 pKa = 5.76YY155 pKa = 9.41TGTARR160 pKa = 11.84PNSWIQDD167 pKa = 3.49VGSRR171 pKa = 11.84AVVEE175 pKa = 4.84LLPSSPPEE183 pKa = 3.48RR184 pKa = 11.84HH185 pKa = 5.68LRR187 pKa = 11.84QAILHH192 pKa = 5.75GSDD195 pKa = 3.51RR196 pKa = 11.84AVEE199 pKa = 3.91LAKK202 pKa = 10.64LDD204 pKa = 4.01LAKK207 pKa = 10.66LQAEE211 pKa = 4.86FEE213 pKa = 4.76SSGHH217 pKa = 6.29PALAAHH223 pKa = 6.47PTDD226 pKa = 3.63VSLDD230 pKa = 3.93TIDD233 pKa = 4.01SVSSFARR240 pKa = 11.84FVLQTTNRR248 pKa = 11.84YY249 pKa = 8.86IPPALQRR256 pKa = 11.84DD257 pKa = 3.97SGRR260 pKa = 11.84SFLDD264 pKa = 3.39SVSDD268 pKa = 3.41ALTAVFEE275 pKa = 4.5HH276 pKa = 6.65PVVCRR281 pKa = 11.84FASTVALNDD290 pKa = 3.56ALRR293 pKa = 11.84FLCRR297 pKa = 11.84RR298 pKa = 11.84NHH300 pKa = 6.91AEE302 pKa = 3.89AADD305 pKa = 3.76AAHH308 pKa = 7.14RR309 pKa = 11.84LYY311 pKa = 10.75HH312 pKa = 6.12AAKK315 pKa = 10.17ALHH318 pKa = 6.29LVLGISTFNILFDD331 pKa = 3.64SAIRR335 pKa = 11.84TGNAKK340 pKa = 10.02RR341 pKa = 11.84AVFFLRR347 pKa = 11.84EE348 pKa = 4.0MQRR351 pKa = 11.84EE352 pKa = 4.42SICPNSHH359 pKa = 4.79TWTLLFSIPSISDD372 pKa = 2.88KK373 pKa = 10.78TALLRR378 pKa = 11.84LIMEE382 pKa = 4.78SGIPMNRR389 pKa = 11.84QGFAIITRR397 pKa = 11.84AAIADD402 pKa = 4.36LLPNQRR408 pKa = 11.84PSCEE412 pKa = 4.32DD413 pKa = 3.34LQQLFADD420 pKa = 4.73LDD422 pKa = 4.51HH423 pKa = 7.25IFGPDD428 pKa = 2.93WLTVDD433 pKa = 4.57SYY435 pKa = 11.99HH436 pKa = 7.21LLLRR440 pKa = 11.84LSSLNPRR447 pKa = 11.84EE448 pKa = 4.2GVVGLLNEE456 pKa = 4.24LPSRR460 pKa = 11.84HH461 pKa = 5.57GLKK464 pKa = 10.17LAPDD468 pKa = 3.82SVRR471 pKa = 11.84YY472 pKa = 8.7LAALNRR478 pKa = 11.84KK479 pKa = 8.47SAITRR484 pKa = 11.84DD485 pKa = 3.68RR486 pKa = 11.84ISDD489 pKa = 3.66FVAAVGAPTCARR501 pKa = 11.84QRR503 pKa = 11.84LVVPRR508 pKa = 11.84FFMAAWRR515 pKa = 11.84RR516 pKa = 11.84CFYY519 pKa = 10.65NVCAVLWSFAATRR532 pKa = 11.84GLITQTMLRR541 pKa = 11.84IVSRR545 pKa = 11.84YY546 pKa = 7.93LVRR549 pKa = 11.84GGAHH553 pKa = 6.57PSSRR557 pKa = 11.84SASRR561 pKa = 11.84KK562 pKa = 9.46RR563 pKa = 11.84IGARR567 pKa = 11.84IVAGVQWRR575 pKa = 11.84RR576 pKa = 11.84GPHH579 pKa = 4.68QVAVMADD586 pKa = 3.76VFPKK590 pKa = 9.76LARR593 pKa = 11.84RR594 pKa = 11.84FGEE597 pKa = 4.07HH598 pKa = 5.17NNARR602 pKa = 11.84RR603 pKa = 11.84WLTGFTADD611 pKa = 4.87DD612 pKa = 3.89GTRR615 pKa = 11.84DD616 pKa = 3.28EE617 pKa = 4.33QMNLFFTTLHH627 pKa = 7.28DD628 pKa = 6.39DD629 pKa = 3.56LGAWKK634 pKa = 10.19RR635 pKa = 11.84FRR637 pKa = 11.84PMSYY641 pKa = 10.96GSLQRR646 pKa = 11.84MLDD649 pKa = 3.25QAYY652 pKa = 9.41EE653 pKa = 3.94ADD655 pKa = 3.34QRR657 pKa = 11.84SISEE661 pKa = 4.23GTLDD665 pKa = 3.57AQGEE669 pKa = 4.54HH670 pKa = 6.56AVSQAVDD677 pKa = 3.33RR678 pKa = 11.84AICVPLTKK686 pKa = 10.56LVDD689 pKa = 4.16GEE691 pKa = 4.8ANWAVQAKK699 pKa = 9.3INQRR703 pKa = 11.84DD704 pKa = 3.38RR705 pKa = 11.84VMDD708 pKa = 4.5AARR711 pKa = 11.84LSRR714 pKa = 11.84RR715 pKa = 11.84RR716 pKa = 11.84GPASGTLHH724 pKa = 7.56DD725 pKa = 4.85LRR727 pKa = 6.49
MM1 pKa = 7.96PSTADD6 pKa = 3.45GDD8 pKa = 4.32CSQTHH13 pKa = 6.0AQTHH17 pKa = 5.2AQTRR21 pKa = 11.84VQTRR25 pKa = 11.84TSAASPNDD33 pKa = 4.06SIRR36 pKa = 11.84PPAPARR42 pKa = 11.84CSRR45 pKa = 11.84MVSRR49 pKa = 11.84CGAAEE54 pKa = 3.88RR55 pKa = 11.84TRR57 pKa = 11.84LQDD60 pKa = 3.26VVQQRR65 pKa = 11.84IARR68 pKa = 11.84LPHH71 pKa = 5.52AQQLARR77 pKa = 11.84LHH79 pKa = 6.44PKK81 pKa = 9.44RR82 pKa = 11.84AAARR86 pKa = 11.84TSSANASQPRR96 pKa = 11.84TQTPAEE102 pKa = 4.38VKK104 pKa = 9.76SQRR107 pKa = 11.84PEE109 pKa = 3.34ARR111 pKa = 11.84QTGRR115 pKa = 11.84SPRR118 pKa = 11.84EE119 pKa = 3.39EE120 pKa = 3.7WQKK123 pKa = 10.76FIHH126 pKa = 6.54RR127 pKa = 11.84LSALPRR133 pKa = 11.84TRR135 pKa = 11.84PSRR138 pKa = 11.84EE139 pKa = 3.83SNGRR143 pKa = 11.84CVEE146 pKa = 3.92LTTAFVHH153 pKa = 6.76HH154 pKa = 5.76YY155 pKa = 9.41TGTARR160 pKa = 11.84PNSWIQDD167 pKa = 3.49VGSRR171 pKa = 11.84AVVEE175 pKa = 4.84LLPSSPPEE183 pKa = 3.48RR184 pKa = 11.84HH185 pKa = 5.68LRR187 pKa = 11.84QAILHH192 pKa = 5.75GSDD195 pKa = 3.51RR196 pKa = 11.84AVEE199 pKa = 3.91LAKK202 pKa = 10.64LDD204 pKa = 4.01LAKK207 pKa = 10.66LQAEE211 pKa = 4.86FEE213 pKa = 4.76SSGHH217 pKa = 6.29PALAAHH223 pKa = 6.47PTDD226 pKa = 3.63VSLDD230 pKa = 3.93TIDD233 pKa = 4.01SVSSFARR240 pKa = 11.84FVLQTTNRR248 pKa = 11.84YY249 pKa = 8.86IPPALQRR256 pKa = 11.84DD257 pKa = 3.97SGRR260 pKa = 11.84SFLDD264 pKa = 3.39SVSDD268 pKa = 3.41ALTAVFEE275 pKa = 4.5HH276 pKa = 6.65PVVCRR281 pKa = 11.84FASTVALNDD290 pKa = 3.56ALRR293 pKa = 11.84FLCRR297 pKa = 11.84RR298 pKa = 11.84NHH300 pKa = 6.91AEE302 pKa = 3.89AADD305 pKa = 3.76AAHH308 pKa = 7.14RR309 pKa = 11.84LYY311 pKa = 10.75HH312 pKa = 6.12AAKK315 pKa = 10.17ALHH318 pKa = 6.29LVLGISTFNILFDD331 pKa = 3.64SAIRR335 pKa = 11.84TGNAKK340 pKa = 10.02RR341 pKa = 11.84AVFFLRR347 pKa = 11.84EE348 pKa = 4.0MQRR351 pKa = 11.84EE352 pKa = 4.42SICPNSHH359 pKa = 4.79TWTLLFSIPSISDD372 pKa = 2.88KK373 pKa = 10.78TALLRR378 pKa = 11.84LIMEE382 pKa = 4.78SGIPMNRR389 pKa = 11.84QGFAIITRR397 pKa = 11.84AAIADD402 pKa = 4.36LLPNQRR408 pKa = 11.84PSCEE412 pKa = 4.32DD413 pKa = 3.34LQQLFADD420 pKa = 4.73LDD422 pKa = 4.51HH423 pKa = 7.25IFGPDD428 pKa = 2.93WLTVDD433 pKa = 4.57SYY435 pKa = 11.99HH436 pKa = 7.21LLLRR440 pKa = 11.84LSSLNPRR447 pKa = 11.84EE448 pKa = 4.2GVVGLLNEE456 pKa = 4.24LPSRR460 pKa = 11.84HH461 pKa = 5.57GLKK464 pKa = 10.17LAPDD468 pKa = 3.82SVRR471 pKa = 11.84YY472 pKa = 8.7LAALNRR478 pKa = 11.84KK479 pKa = 8.47SAITRR484 pKa = 11.84DD485 pKa = 3.68RR486 pKa = 11.84ISDD489 pKa = 3.66FVAAVGAPTCARR501 pKa = 11.84QRR503 pKa = 11.84LVVPRR508 pKa = 11.84FFMAAWRR515 pKa = 11.84RR516 pKa = 11.84CFYY519 pKa = 10.65NVCAVLWSFAATRR532 pKa = 11.84GLITQTMLRR541 pKa = 11.84IVSRR545 pKa = 11.84YY546 pKa = 7.93LVRR549 pKa = 11.84GGAHH553 pKa = 6.57PSSRR557 pKa = 11.84SASRR561 pKa = 11.84KK562 pKa = 9.46RR563 pKa = 11.84IGARR567 pKa = 11.84IVAGVQWRR575 pKa = 11.84RR576 pKa = 11.84GPHH579 pKa = 4.68QVAVMADD586 pKa = 3.76VFPKK590 pKa = 9.76LARR593 pKa = 11.84RR594 pKa = 11.84FGEE597 pKa = 4.07HH598 pKa = 5.17NNARR602 pKa = 11.84RR603 pKa = 11.84WLTGFTADD611 pKa = 4.87DD612 pKa = 3.89GTRR615 pKa = 11.84DD616 pKa = 3.28EE617 pKa = 4.33QMNLFFTTLHH627 pKa = 7.28DD628 pKa = 6.39DD629 pKa = 3.56LGAWKK634 pKa = 10.19RR635 pKa = 11.84FRR637 pKa = 11.84PMSYY641 pKa = 10.96GSLQRR646 pKa = 11.84MLDD649 pKa = 3.25QAYY652 pKa = 9.41EE653 pKa = 3.94ADD655 pKa = 3.34QRR657 pKa = 11.84SISEE661 pKa = 4.23GTLDD665 pKa = 3.57AQGEE669 pKa = 4.54HH670 pKa = 6.56AVSQAVDD677 pKa = 3.33RR678 pKa = 11.84AICVPLTKK686 pKa = 10.56LVDD689 pKa = 4.16GEE691 pKa = 4.8ANWAVQAKK699 pKa = 9.3INQRR703 pKa = 11.84DD704 pKa = 3.38RR705 pKa = 11.84VMDD708 pKa = 4.5AARR711 pKa = 11.84LSRR714 pKa = 11.84RR715 pKa = 11.84RR716 pKa = 11.84GPASGTLHH724 pKa = 7.56DD725 pKa = 4.85LRR727 pKa = 6.49
Molecular weight: 81.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3461870 |
48 |
5823 |
604.6 |
66.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.704 ± 0.028 | 1.065 ± 0.009 |
5.979 ± 0.021 | 5.86 ± 0.021 |
3.552 ± 0.018 | 6.723 ± 0.023 |
2.517 ± 0.013 | 4.695 ± 0.019 |
4.814 ± 0.022 | 9.142 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.987 ± 0.011 | 3.41 ± 0.015 |
5.927 ± 0.028 | 4.2 ± 0.019 |
6.264 ± 0.021 | 8.13 ± 0.029 |
5.783 ± 0.018 | 6.21 ± 0.019 |
1.365 ± 0.009 | 2.674 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
