
Siansivirga zeaxanthinifaciens CC-SAMT-1
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Siansivirga; Siansivirga zeaxanthinifaciens
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
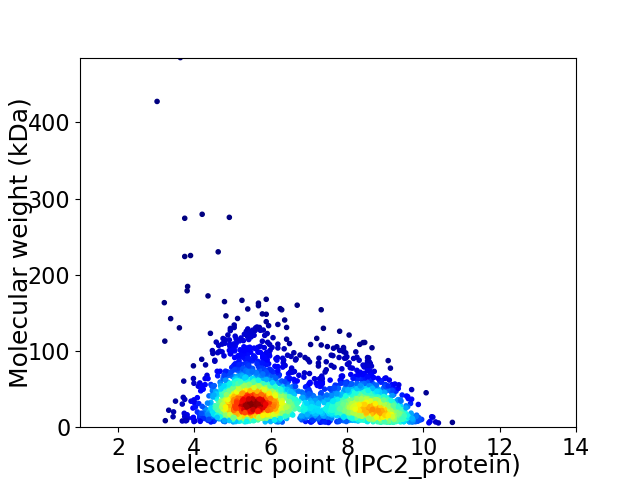
Virtual 2D-PAGE plot for 2744 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
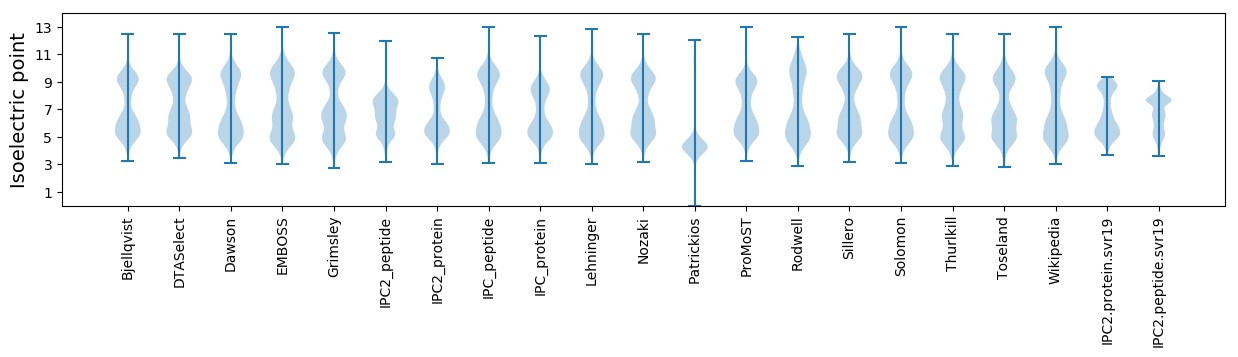
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5WCM4|A0A0C5WCM4_9FLAO Uncharacterized protein OS=Siansivirga zeaxanthinifaciens CC-SAMT-1 OX=1454006 GN=AW14_03220 PE=4 SV=1
MM1 pKa = 7.11GHH3 pKa = 5.59NTGALTEE10 pKa = 4.21QTAEE14 pKa = 4.4VPASLPGAKK23 pKa = 9.56RR24 pKa = 11.84IGRR27 pKa = 11.84EE28 pKa = 3.65WLIKK32 pKa = 9.51RR33 pKa = 11.84TGDD36 pKa = 3.15VGTVDD41 pKa = 4.93LSFDD45 pKa = 3.67VTGLTLSGSLFIDD58 pKa = 4.51FKK60 pKa = 11.62LLIDD64 pKa = 4.14ADD66 pKa = 4.35GDD68 pKa = 3.97FSSGATIINASSFAGGKK85 pKa = 9.87VSFTGVTLPDD95 pKa = 3.15ANYY98 pKa = 8.03ITFVTDD104 pKa = 3.18VPLFLSSNLWLKK116 pKa = 10.58GNQGVTNSGSNLAGWVDD133 pKa = 3.38QTGINTFTVSGNPQTGVSKK152 pKa = 10.91INFNNAIDD160 pKa = 3.86FDD162 pKa = 4.35GAGDD166 pKa = 4.05YY167 pKa = 9.24LTGNAAITFQNVYY180 pKa = 10.91AVIKK184 pKa = 10.56RR185 pKa = 11.84EE186 pKa = 4.01NTGDD190 pKa = 3.83LTVLSASSGSANRR203 pKa = 11.84GYY205 pKa = 11.33VMKK208 pKa = 10.22SNNMSTGNNDD218 pKa = 2.47GSNYY222 pKa = 8.6FRR224 pKa = 11.84SVGTLGTDD232 pKa = 3.31KK233 pKa = 11.18ARR235 pKa = 11.84IGHH238 pKa = 4.96VEE240 pKa = 3.92IVAGQPATNQKK251 pKa = 10.76SYY253 pKa = 11.33IDD255 pKa = 3.66GQQFNTVSLVGSGNFVNYY273 pKa = 10.42SDD275 pKa = 3.76VPYY278 pKa = 10.69VGRR281 pKa = 11.84SQDD284 pKa = 3.39NAQPDD289 pKa = 4.04YY290 pKa = 11.18FNGKK294 pKa = 7.51IAEE297 pKa = 4.43IIMYY301 pKa = 8.35PAAHH305 pKa = 6.79TSRR308 pKa = 11.84EE309 pKa = 3.71RR310 pKa = 11.84SKK312 pKa = 10.39IQSYY316 pKa = 9.87LAIKK320 pKa = 10.02YY321 pKa = 8.94GISLDD326 pKa = 4.02PSASAYY332 pKa = 10.44LSSSEE337 pKa = 3.87ALIWTDD343 pKa = 2.95LTYY346 pKa = 10.71WNDD349 pKa = 2.87VFGIGKK355 pKa = 9.82DD356 pKa = 3.57DD357 pKa = 4.28ASSLNQTQSNSINTGSGDD375 pKa = 3.48GTGQSGKK382 pKa = 11.05GNIIISNPSSLNNGDD397 pKa = 3.9FLMIGHH403 pKa = 7.51DD404 pKa = 3.55NGALTEE410 pKa = 4.09QATDD414 pKa = 3.6LPTNYY419 pKa = 9.55IGAARR424 pKa = 11.84LARR427 pKa = 11.84EE428 pKa = 4.44WKK430 pKa = 9.79VKK432 pKa = 8.94HH433 pKa = 6.03TGDD436 pKa = 4.05VGTVDD441 pKa = 5.21LKK443 pKa = 10.86IDD445 pKa = 3.65LTGLNLSGVPLNNFQLLLDD464 pKa = 4.23ADD466 pKa = 4.53GDD468 pKa = 4.15FSSGATAVIPASFSSNILTFTSISLIDD495 pKa = 3.53GTVFALVTKK504 pKa = 9.32TAGPGVAGASLWLKK518 pKa = 9.56TDD520 pKa = 3.82DD521 pKa = 5.39GITASGSNLTGWTDD535 pKa = 3.27QTGTNTFSLNGTPTVVNNSVNFNPIVNFTNPTSPGVPVNGLIGDD579 pKa = 4.01TEE581 pKa = 4.18ISGVEE586 pKa = 3.56AFGVFKK592 pKa = 11.27YY593 pKa = 10.74KK594 pKa = 10.72DD595 pKa = 3.25IVNQGTIVGNTTGDD609 pKa = 3.29GGFFAGLLNVMKK621 pKa = 10.92NRR623 pKa = 11.84DD624 pKa = 3.7TNGNDD629 pKa = 3.09QTFANANLSTVFSISNIDD647 pKa = 3.84LSPTLAQATVNGLAASVTQEE667 pKa = 3.96KK668 pKa = 10.69GVDD671 pKa = 3.99FASVNLTPAIGGGANVGGLNGDD693 pKa = 3.68LAEE696 pKa = 4.12LVVYY700 pKa = 10.4SKK702 pKa = 11.13SLSALEE708 pKa = 4.0KK709 pKa = 10.77QKK711 pKa = 10.44VQSYY715 pKa = 9.68LAVKK719 pKa = 10.27YY720 pKa = 10.44GITLDD725 pKa = 3.7QSISSYY731 pKa = 10.54VSSNGTVIWDD741 pKa = 3.48NTSYY745 pKa = 10.64WNDD748 pKa = 2.68VFGIGRR754 pKa = 11.84EE755 pKa = 4.11DD756 pKa = 3.76ASGLNQTQSNSINSGSGDD774 pKa = 3.41GTGQSGKK781 pKa = 10.96GNIVISNPSSLEE793 pKa = 4.2DD794 pKa = 4.15GDD796 pKa = 4.36FLLIGHH802 pKa = 7.28DD803 pKa = 3.51NGALTEE809 pKa = 4.13QVTDD813 pKa = 4.7LPADD817 pKa = 3.52YY818 pKa = 10.82AGRR821 pKa = 11.84TRR823 pKa = 11.84LTRR826 pKa = 11.84EE827 pKa = 3.93WKK829 pKa = 9.95VKK831 pKa = 8.86HH832 pKa = 6.03TGDD835 pKa = 4.05VGTVDD840 pKa = 5.21LKK842 pKa = 10.86IDD844 pKa = 3.65LTGLNLSGVPLNNFQLLLDD863 pKa = 4.23ADD865 pKa = 4.53GDD867 pKa = 4.15FSSGATAVIPASFSSNILTFTGISLTDD894 pKa = 3.31GTVFALVTKK903 pKa = 9.32TAGPGVAGASLWLKK917 pKa = 9.41TDD919 pKa = 3.36AGLTSSGSNLTGWTDD934 pKa = 3.28QVGINTFNVNGSPEE948 pKa = 4.16VVDD951 pKa = 4.08NNINFNPSVYY961 pKa = 9.87FANAPGSGLPEE972 pKa = 5.17IGLIGNTEE980 pKa = 4.15IIGVEE985 pKa = 3.54AFGIFRR991 pKa = 11.84HH992 pKa = 6.26ADD994 pKa = 3.3LVNQGTILGNSTGEE1008 pKa = 4.05GGFFSSTLGIISTGDD1023 pKa = 3.32SNGNDD1028 pKa = 2.99QTFEE1032 pKa = 4.03NPNVGLEE1039 pKa = 3.85FSIVTIDD1046 pKa = 4.05VSPTLADD1053 pKa = 3.56AKK1055 pKa = 10.52IDD1057 pKa = 3.65GLAASVTQDD1066 pKa = 2.65SGTDD1070 pKa = 3.72FSSVSLIPVIGGGSLMTKK1088 pKa = 10.58LNGEE1092 pKa = 4.22LVEE1095 pKa = 4.77LIVYY1099 pKa = 8.4PSSLSAIDD1107 pKa = 3.4KK1108 pKa = 10.67RR1109 pKa = 11.84KK1110 pKa = 8.01VQSYY1114 pKa = 9.66LAIKK1118 pKa = 10.06YY1119 pKa = 8.97GISLDD1124 pKa = 3.83QSVTSYY1130 pKa = 9.68MSSSGTVIWNNTTYY1144 pKa = 10.31WNDD1147 pKa = 2.77VFGIGKK1153 pKa = 9.94DD1154 pKa = 3.57DD1155 pKa = 3.73VSGLNQSQSNSINTGSGDD1173 pKa = 3.52GTGQTGKK1180 pKa = 11.13GNIVISNPSSLEE1192 pKa = 4.21DD1193 pKa = 4.02GDD1195 pKa = 4.36FLMIGHH1201 pKa = 7.47DD1202 pKa = 3.37NGALVEE1208 pKa = 4.14QSTDD1212 pKa = 3.23LPTSLSCFSRR1222 pKa = 11.84IRR1224 pKa = 11.84RR1225 pKa = 11.84EE1226 pKa = 3.71WKK1228 pKa = 9.8VNHH1231 pKa = 6.35TNDD1234 pKa = 3.12VGTVDD1239 pKa = 5.01LSFDD1243 pKa = 3.44IKK1245 pKa = 10.96GLTVGGTLKK1254 pKa = 10.75EE1255 pKa = 4.11DD1256 pKa = 3.24FRR1258 pKa = 11.84LVIDD1262 pKa = 3.94QDD1264 pKa = 3.37GDD1266 pKa = 4.17GNFTTGTVTVITPNSLTSGVLLFQGVTLNSGAVFTFVTGPEE1307 pKa = 4.45TISPTITCVSDD1318 pKa = 3.26LTQPSDD1324 pKa = 3.68AGNCSAVVNNIAPTASDD1341 pKa = 3.13NCAVTTQTWTLTGATVKK1358 pKa = 10.8NSATTGINDD1367 pKa = 3.81ASGEE1371 pKa = 4.28TFNLGVTTVTYY1382 pKa = 9.24TVSDD1386 pKa = 3.39AAGNTNSCSFDD1397 pKa = 3.25VTITDD1402 pKa = 3.78TEE1404 pKa = 4.54LPTIATASNITVSSDD1419 pKa = 2.75TGTCTYY1425 pKa = 10.92ASSQLTPPTTDD1436 pKa = 3.77DD1437 pKa = 3.51NCGVASVVASPSSLAIGTNTVTWTVTDD1464 pKa = 3.57NSGNTATTTQTVTVEE1479 pKa = 4.03DD1480 pKa = 4.22TEE1482 pKa = 4.78LPTIATASNITVSSDD1497 pKa = 2.75TGTCTYY1503 pKa = 10.92ASSQLTPPTVNDD1515 pKa = 3.34NCGVASVVASPSSLAIGTNTVTWTVTDD1542 pKa = 3.57NSGNTATTTQTVTVEE1557 pKa = 4.22DD1558 pKa = 3.87NQLPTIATVGAVTVPADD1575 pKa = 3.88AGVCTYY1581 pKa = 9.18STSQLPPPSTDD1592 pKa = 3.54DD1593 pKa = 3.51NCGVASVVVSPSSLALGAHH1612 pKa = 5.18TVTWTVTDD1620 pKa = 3.57NSGNTATTTEE1630 pKa = 4.4TVTVVDD1636 pKa = 4.19TEE1638 pKa = 4.57LPTIATVGAVTVPADD1653 pKa = 3.88AGVCTYY1659 pKa = 9.18STSQLPPPSTDD1670 pKa = 3.54DD1671 pKa = 3.51NCGVASVVGIPSTLNAGSNTVTWIVVDD1698 pKa = 4.9DD1699 pKa = 4.32SGNQNRR1705 pKa = 11.84SIQTVIVVEE1714 pKa = 4.24NEE1716 pKa = 3.71LPTISNLGPITVNSDD1731 pKa = 3.29PGVCTYY1737 pKa = 11.2ASSQLTPPTTNDD1749 pKa = 2.9NCGVASVVASPSSLVLGDD1767 pKa = 5.01NLVTWTVTDD1776 pKa = 3.43NAGNVANSLQTVTVVDD1792 pKa = 3.76NEE1794 pKa = 4.16RR1795 pKa = 11.84PTIVIAGPVTVSSDD1809 pKa = 3.43PGVCTYY1815 pKa = 11.2ASSQLTPPTTNDD1827 pKa = 2.9NCGVASVVVSPSSLAIGNNTVTWTVTDD1854 pKa = 4.05DD1855 pKa = 3.75SGNTATATQIVTVEE1869 pKa = 4.12DD1870 pKa = 4.04TEE1872 pKa = 4.46NPTIASVGSVTVNVDD1887 pKa = 3.5AGLCTYY1893 pKa = 7.63DD1894 pKa = 3.82TSQLPPPAADD1904 pKa = 4.17DD1905 pKa = 3.72NCGVLFVISDD1915 pKa = 3.65KK1916 pKa = 9.14TTLNVGVNTLTWYY1929 pKa = 10.69AFDD1932 pKa = 4.23FEE1934 pKa = 6.33LNFSSTTQTVIVVEE1948 pKa = 4.39NEE1950 pKa = 3.71APTIATLGPITVSSDD1965 pKa = 3.31LGVCTYY1971 pKa = 10.96DD1972 pKa = 4.13SSQLPPPATSDD1983 pKa = 3.34NCGVASVVASPSTLVLGPNTVTWTVTDD2010 pKa = 3.55NAGNIANSIQTVTVEE2025 pKa = 4.49DD2026 pKa = 3.85NQAPTIATVSAVTVSADD2043 pKa = 4.19AGVCTYY2049 pKa = 8.07NTSQLPPPATSDD2061 pKa = 3.34NCGVASVVASPSTLVLGSNTVTWTVTDD2088 pKa = 3.33NAGNIKK2094 pKa = 7.79TTIQGVTVQDD2104 pKa = 3.69NEE2106 pKa = 4.35APTFTSVSAITVSADD2121 pKa = 3.33AGVCTYY2127 pKa = 11.08ASSQLTPPSVTDD2139 pKa = 3.26NCGVAGILRR2148 pKa = 11.84NPGVLSVGINTVTWTAIDD2166 pKa = 3.7DD2167 pKa = 4.03SGNQKK2172 pKa = 10.75SIIQTVTVEE2181 pKa = 4.23DD2182 pKa = 4.26NEE2184 pKa = 4.69DD2185 pKa = 3.4PTIASASNITVSSDD2199 pKa = 3.27AGTCTYY2205 pKa = 11.02ASSQLTPPTVNDD2217 pKa = 3.34NCGVASVVASPSTLVLGPNTVTWTVTDD2244 pKa = 3.63NAGNTASTTQTVTVEE2259 pKa = 4.21DD2260 pKa = 4.36NEE2262 pKa = 4.72DD2263 pKa = 3.42PTIAAASNITVSSDD2277 pKa = 3.27AGTCTYY2283 pKa = 11.35ASAQLTPPTVNDD2295 pKa = 3.34NCGVASVVASPSTIVLGPNTVTWTVTDD2322 pKa = 3.63NAGNTASTTQTVTVEE2337 pKa = 4.21DD2338 pKa = 4.36NEE2340 pKa = 4.69DD2341 pKa = 3.4PTIASASNITVSSDD2355 pKa = 3.27AGTCTYY2361 pKa = 11.02ASSQLTPPTVNDD2373 pKa = 3.34NCGVASVVASPSSLALGANTVTWTVTDD2400 pKa = 3.63NAGNTASTTQTVTVEE2415 pKa = 4.21DD2416 pKa = 4.36NEE2418 pKa = 4.72DD2419 pKa = 3.42PTIAAASNITVSSDD2433 pKa = 3.27AGTCTYY2439 pKa = 11.35ASAQLTPPTVNDD2451 pKa = 3.34NCGVASVVASPSSLALGANTVTWTVTDD2478 pKa = 3.63NAGNTASTTQTVTVEE2493 pKa = 4.2DD2494 pKa = 3.84NQLPTVSCPANQTVVADD2511 pKa = 3.31VSGFYY2516 pKa = 10.01TLPDD2520 pKa = 3.58YY2521 pKa = 11.03FGTGAGSATDD2531 pKa = 3.63TCSGTIAVYY2540 pKa = 10.21SQSPTVGTQLGIGVYY2555 pKa = 9.79TITLTAEE2562 pKa = 4.26DD2563 pKa = 3.76ASGNIGSCDD2572 pKa = 3.53FEE2574 pKa = 4.66LTVSAPLGLEE2584 pKa = 3.91DD2585 pKa = 5.21LNRR2588 pKa = 11.84DD2589 pKa = 2.81IKK2591 pKa = 10.14TIQLYY2596 pKa = 9.65PNPSSNLVYY2605 pKa = 10.52LSNPSRR2611 pKa = 11.84INLQEE2616 pKa = 3.62VSIYY2620 pKa = 10.95DD2621 pKa = 3.32VTGRR2625 pKa = 11.84LIEE2628 pKa = 4.21TVKK2631 pKa = 10.83LKK2633 pKa = 10.85SEE2635 pKa = 4.24EE2636 pKa = 4.03KK2637 pKa = 10.58QEE2639 pKa = 4.28VIDD2642 pKa = 3.81MSHH2645 pKa = 7.08WDD2647 pKa = 3.05SAMYY2651 pKa = 10.61LFVIKK2656 pKa = 10.44SDD2658 pKa = 3.49LGQVTKK2664 pKa = 10.9SVVKK2668 pKa = 10.13EE2669 pKa = 3.84
MM1 pKa = 7.11GHH3 pKa = 5.59NTGALTEE10 pKa = 4.21QTAEE14 pKa = 4.4VPASLPGAKK23 pKa = 9.56RR24 pKa = 11.84IGRR27 pKa = 11.84EE28 pKa = 3.65WLIKK32 pKa = 9.51RR33 pKa = 11.84TGDD36 pKa = 3.15VGTVDD41 pKa = 4.93LSFDD45 pKa = 3.67VTGLTLSGSLFIDD58 pKa = 4.51FKK60 pKa = 11.62LLIDD64 pKa = 4.14ADD66 pKa = 4.35GDD68 pKa = 3.97FSSGATIINASSFAGGKK85 pKa = 9.87VSFTGVTLPDD95 pKa = 3.15ANYY98 pKa = 8.03ITFVTDD104 pKa = 3.18VPLFLSSNLWLKK116 pKa = 10.58GNQGVTNSGSNLAGWVDD133 pKa = 3.38QTGINTFTVSGNPQTGVSKK152 pKa = 10.91INFNNAIDD160 pKa = 3.86FDD162 pKa = 4.35GAGDD166 pKa = 4.05YY167 pKa = 9.24LTGNAAITFQNVYY180 pKa = 10.91AVIKK184 pKa = 10.56RR185 pKa = 11.84EE186 pKa = 4.01NTGDD190 pKa = 3.83LTVLSASSGSANRR203 pKa = 11.84GYY205 pKa = 11.33VMKK208 pKa = 10.22SNNMSTGNNDD218 pKa = 2.47GSNYY222 pKa = 8.6FRR224 pKa = 11.84SVGTLGTDD232 pKa = 3.31KK233 pKa = 11.18ARR235 pKa = 11.84IGHH238 pKa = 4.96VEE240 pKa = 3.92IVAGQPATNQKK251 pKa = 10.76SYY253 pKa = 11.33IDD255 pKa = 3.66GQQFNTVSLVGSGNFVNYY273 pKa = 10.42SDD275 pKa = 3.76VPYY278 pKa = 10.69VGRR281 pKa = 11.84SQDD284 pKa = 3.39NAQPDD289 pKa = 4.04YY290 pKa = 11.18FNGKK294 pKa = 7.51IAEE297 pKa = 4.43IIMYY301 pKa = 8.35PAAHH305 pKa = 6.79TSRR308 pKa = 11.84EE309 pKa = 3.71RR310 pKa = 11.84SKK312 pKa = 10.39IQSYY316 pKa = 9.87LAIKK320 pKa = 10.02YY321 pKa = 8.94GISLDD326 pKa = 4.02PSASAYY332 pKa = 10.44LSSSEE337 pKa = 3.87ALIWTDD343 pKa = 2.95LTYY346 pKa = 10.71WNDD349 pKa = 2.87VFGIGKK355 pKa = 9.82DD356 pKa = 3.57DD357 pKa = 4.28ASSLNQTQSNSINTGSGDD375 pKa = 3.48GTGQSGKK382 pKa = 11.05GNIIISNPSSLNNGDD397 pKa = 3.9FLMIGHH403 pKa = 7.51DD404 pKa = 3.55NGALTEE410 pKa = 4.09QATDD414 pKa = 3.6LPTNYY419 pKa = 9.55IGAARR424 pKa = 11.84LARR427 pKa = 11.84EE428 pKa = 4.44WKK430 pKa = 9.79VKK432 pKa = 8.94HH433 pKa = 6.03TGDD436 pKa = 4.05VGTVDD441 pKa = 5.21LKK443 pKa = 10.86IDD445 pKa = 3.65LTGLNLSGVPLNNFQLLLDD464 pKa = 4.23ADD466 pKa = 4.53GDD468 pKa = 4.15FSSGATAVIPASFSSNILTFTSISLIDD495 pKa = 3.53GTVFALVTKK504 pKa = 9.32TAGPGVAGASLWLKK518 pKa = 9.56TDD520 pKa = 3.82DD521 pKa = 5.39GITASGSNLTGWTDD535 pKa = 3.27QTGTNTFSLNGTPTVVNNSVNFNPIVNFTNPTSPGVPVNGLIGDD579 pKa = 4.01TEE581 pKa = 4.18ISGVEE586 pKa = 3.56AFGVFKK592 pKa = 11.27YY593 pKa = 10.74KK594 pKa = 10.72DD595 pKa = 3.25IVNQGTIVGNTTGDD609 pKa = 3.29GGFFAGLLNVMKK621 pKa = 10.92NRR623 pKa = 11.84DD624 pKa = 3.7TNGNDD629 pKa = 3.09QTFANANLSTVFSISNIDD647 pKa = 3.84LSPTLAQATVNGLAASVTQEE667 pKa = 3.96KK668 pKa = 10.69GVDD671 pKa = 3.99FASVNLTPAIGGGANVGGLNGDD693 pKa = 3.68LAEE696 pKa = 4.12LVVYY700 pKa = 10.4SKK702 pKa = 11.13SLSALEE708 pKa = 4.0KK709 pKa = 10.77QKK711 pKa = 10.44VQSYY715 pKa = 9.68LAVKK719 pKa = 10.27YY720 pKa = 10.44GITLDD725 pKa = 3.7QSISSYY731 pKa = 10.54VSSNGTVIWDD741 pKa = 3.48NTSYY745 pKa = 10.64WNDD748 pKa = 2.68VFGIGRR754 pKa = 11.84EE755 pKa = 4.11DD756 pKa = 3.76ASGLNQTQSNSINSGSGDD774 pKa = 3.41GTGQSGKK781 pKa = 10.96GNIVISNPSSLEE793 pKa = 4.2DD794 pKa = 4.15GDD796 pKa = 4.36FLLIGHH802 pKa = 7.28DD803 pKa = 3.51NGALTEE809 pKa = 4.13QVTDD813 pKa = 4.7LPADD817 pKa = 3.52YY818 pKa = 10.82AGRR821 pKa = 11.84TRR823 pKa = 11.84LTRR826 pKa = 11.84EE827 pKa = 3.93WKK829 pKa = 9.95VKK831 pKa = 8.86HH832 pKa = 6.03TGDD835 pKa = 4.05VGTVDD840 pKa = 5.21LKK842 pKa = 10.86IDD844 pKa = 3.65LTGLNLSGVPLNNFQLLLDD863 pKa = 4.23ADD865 pKa = 4.53GDD867 pKa = 4.15FSSGATAVIPASFSSNILTFTGISLTDD894 pKa = 3.31GTVFALVTKK903 pKa = 9.32TAGPGVAGASLWLKK917 pKa = 9.41TDD919 pKa = 3.36AGLTSSGSNLTGWTDD934 pKa = 3.28QVGINTFNVNGSPEE948 pKa = 4.16VVDD951 pKa = 4.08NNINFNPSVYY961 pKa = 9.87FANAPGSGLPEE972 pKa = 5.17IGLIGNTEE980 pKa = 4.15IIGVEE985 pKa = 3.54AFGIFRR991 pKa = 11.84HH992 pKa = 6.26ADD994 pKa = 3.3LVNQGTILGNSTGEE1008 pKa = 4.05GGFFSSTLGIISTGDD1023 pKa = 3.32SNGNDD1028 pKa = 2.99QTFEE1032 pKa = 4.03NPNVGLEE1039 pKa = 3.85FSIVTIDD1046 pKa = 4.05VSPTLADD1053 pKa = 3.56AKK1055 pKa = 10.52IDD1057 pKa = 3.65GLAASVTQDD1066 pKa = 2.65SGTDD1070 pKa = 3.72FSSVSLIPVIGGGSLMTKK1088 pKa = 10.58LNGEE1092 pKa = 4.22LVEE1095 pKa = 4.77LIVYY1099 pKa = 8.4PSSLSAIDD1107 pKa = 3.4KK1108 pKa = 10.67RR1109 pKa = 11.84KK1110 pKa = 8.01VQSYY1114 pKa = 9.66LAIKK1118 pKa = 10.06YY1119 pKa = 8.97GISLDD1124 pKa = 3.83QSVTSYY1130 pKa = 9.68MSSSGTVIWNNTTYY1144 pKa = 10.31WNDD1147 pKa = 2.77VFGIGKK1153 pKa = 9.94DD1154 pKa = 3.57DD1155 pKa = 3.73VSGLNQSQSNSINTGSGDD1173 pKa = 3.52GTGQTGKK1180 pKa = 11.13GNIVISNPSSLEE1192 pKa = 4.21DD1193 pKa = 4.02GDD1195 pKa = 4.36FLMIGHH1201 pKa = 7.47DD1202 pKa = 3.37NGALVEE1208 pKa = 4.14QSTDD1212 pKa = 3.23LPTSLSCFSRR1222 pKa = 11.84IRR1224 pKa = 11.84RR1225 pKa = 11.84EE1226 pKa = 3.71WKK1228 pKa = 9.8VNHH1231 pKa = 6.35TNDD1234 pKa = 3.12VGTVDD1239 pKa = 5.01LSFDD1243 pKa = 3.44IKK1245 pKa = 10.96GLTVGGTLKK1254 pKa = 10.75EE1255 pKa = 4.11DD1256 pKa = 3.24FRR1258 pKa = 11.84LVIDD1262 pKa = 3.94QDD1264 pKa = 3.37GDD1266 pKa = 4.17GNFTTGTVTVITPNSLTSGVLLFQGVTLNSGAVFTFVTGPEE1307 pKa = 4.45TISPTITCVSDD1318 pKa = 3.26LTQPSDD1324 pKa = 3.68AGNCSAVVNNIAPTASDD1341 pKa = 3.13NCAVTTQTWTLTGATVKK1358 pKa = 10.8NSATTGINDD1367 pKa = 3.81ASGEE1371 pKa = 4.28TFNLGVTTVTYY1382 pKa = 9.24TVSDD1386 pKa = 3.39AAGNTNSCSFDD1397 pKa = 3.25VTITDD1402 pKa = 3.78TEE1404 pKa = 4.54LPTIATASNITVSSDD1419 pKa = 2.75TGTCTYY1425 pKa = 10.92ASSQLTPPTTDD1436 pKa = 3.77DD1437 pKa = 3.51NCGVASVVASPSSLAIGTNTVTWTVTDD1464 pKa = 3.57NSGNTATTTQTVTVEE1479 pKa = 4.03DD1480 pKa = 4.22TEE1482 pKa = 4.78LPTIATASNITVSSDD1497 pKa = 2.75TGTCTYY1503 pKa = 10.92ASSQLTPPTVNDD1515 pKa = 3.34NCGVASVVASPSSLAIGTNTVTWTVTDD1542 pKa = 3.57NSGNTATTTQTVTVEE1557 pKa = 4.22DD1558 pKa = 3.87NQLPTIATVGAVTVPADD1575 pKa = 3.88AGVCTYY1581 pKa = 9.18STSQLPPPSTDD1592 pKa = 3.54DD1593 pKa = 3.51NCGVASVVVSPSSLALGAHH1612 pKa = 5.18TVTWTVTDD1620 pKa = 3.57NSGNTATTTEE1630 pKa = 4.4TVTVVDD1636 pKa = 4.19TEE1638 pKa = 4.57LPTIATVGAVTVPADD1653 pKa = 3.88AGVCTYY1659 pKa = 9.18STSQLPPPSTDD1670 pKa = 3.54DD1671 pKa = 3.51NCGVASVVGIPSTLNAGSNTVTWIVVDD1698 pKa = 4.9DD1699 pKa = 4.32SGNQNRR1705 pKa = 11.84SIQTVIVVEE1714 pKa = 4.24NEE1716 pKa = 3.71LPTISNLGPITVNSDD1731 pKa = 3.29PGVCTYY1737 pKa = 11.2ASSQLTPPTTNDD1749 pKa = 2.9NCGVASVVASPSSLVLGDD1767 pKa = 5.01NLVTWTVTDD1776 pKa = 3.43NAGNVANSLQTVTVVDD1792 pKa = 3.76NEE1794 pKa = 4.16RR1795 pKa = 11.84PTIVIAGPVTVSSDD1809 pKa = 3.43PGVCTYY1815 pKa = 11.2ASSQLTPPTTNDD1827 pKa = 2.9NCGVASVVVSPSSLAIGNNTVTWTVTDD1854 pKa = 4.05DD1855 pKa = 3.75SGNTATATQIVTVEE1869 pKa = 4.12DD1870 pKa = 4.04TEE1872 pKa = 4.46NPTIASVGSVTVNVDD1887 pKa = 3.5AGLCTYY1893 pKa = 7.63DD1894 pKa = 3.82TSQLPPPAADD1904 pKa = 4.17DD1905 pKa = 3.72NCGVLFVISDD1915 pKa = 3.65KK1916 pKa = 9.14TTLNVGVNTLTWYY1929 pKa = 10.69AFDD1932 pKa = 4.23FEE1934 pKa = 6.33LNFSSTTQTVIVVEE1948 pKa = 4.39NEE1950 pKa = 3.71APTIATLGPITVSSDD1965 pKa = 3.31LGVCTYY1971 pKa = 10.96DD1972 pKa = 4.13SSQLPPPATSDD1983 pKa = 3.34NCGVASVVASPSTLVLGPNTVTWTVTDD2010 pKa = 3.55NAGNIANSIQTVTVEE2025 pKa = 4.49DD2026 pKa = 3.85NQAPTIATVSAVTVSADD2043 pKa = 4.19AGVCTYY2049 pKa = 8.07NTSQLPPPATSDD2061 pKa = 3.34NCGVASVVASPSTLVLGSNTVTWTVTDD2088 pKa = 3.33NAGNIKK2094 pKa = 7.79TTIQGVTVQDD2104 pKa = 3.69NEE2106 pKa = 4.35APTFTSVSAITVSADD2121 pKa = 3.33AGVCTYY2127 pKa = 11.08ASSQLTPPSVTDD2139 pKa = 3.26NCGVAGILRR2148 pKa = 11.84NPGVLSVGINTVTWTAIDD2166 pKa = 3.7DD2167 pKa = 4.03SGNQKK2172 pKa = 10.75SIIQTVTVEE2181 pKa = 4.23DD2182 pKa = 4.26NEE2184 pKa = 4.69DD2185 pKa = 3.4PTIASASNITVSSDD2199 pKa = 3.27AGTCTYY2205 pKa = 11.02ASSQLTPPTVNDD2217 pKa = 3.34NCGVASVVASPSTLVLGPNTVTWTVTDD2244 pKa = 3.63NAGNTASTTQTVTVEE2259 pKa = 4.21DD2260 pKa = 4.36NEE2262 pKa = 4.72DD2263 pKa = 3.42PTIAAASNITVSSDD2277 pKa = 3.27AGTCTYY2283 pKa = 11.35ASAQLTPPTVNDD2295 pKa = 3.34NCGVASVVASPSTIVLGPNTVTWTVTDD2322 pKa = 3.63NAGNTASTTQTVTVEE2337 pKa = 4.21DD2338 pKa = 4.36NEE2340 pKa = 4.69DD2341 pKa = 3.4PTIASASNITVSSDD2355 pKa = 3.27AGTCTYY2361 pKa = 11.02ASSQLTPPTVNDD2373 pKa = 3.34NCGVASVVASPSSLALGANTVTWTVTDD2400 pKa = 3.63NAGNTASTTQTVTVEE2415 pKa = 4.21DD2416 pKa = 4.36NEE2418 pKa = 4.72DD2419 pKa = 3.42PTIAAASNITVSSDD2433 pKa = 3.27AGTCTYY2439 pKa = 11.35ASAQLTPPTVNDD2451 pKa = 3.34NCGVASVVASPSSLALGANTVTWTVTDD2478 pKa = 3.63NAGNTASTTQTVTVEE2493 pKa = 4.2DD2494 pKa = 3.84NQLPTVSCPANQTVVADD2511 pKa = 3.31VSGFYY2516 pKa = 10.01TLPDD2520 pKa = 3.58YY2521 pKa = 11.03FGTGAGSATDD2531 pKa = 3.63TCSGTIAVYY2540 pKa = 10.21SQSPTVGTQLGIGVYY2555 pKa = 9.79TITLTAEE2562 pKa = 4.26DD2563 pKa = 3.76ASGNIGSCDD2572 pKa = 3.53FEE2574 pKa = 4.66LTVSAPLGLEE2584 pKa = 3.91DD2585 pKa = 5.21LNRR2588 pKa = 11.84DD2589 pKa = 2.81IKK2591 pKa = 10.14TIQLYY2596 pKa = 9.65PNPSSNLVYY2605 pKa = 10.52LSNPSRR2611 pKa = 11.84INLQEE2616 pKa = 3.62VSIYY2620 pKa = 10.95DD2621 pKa = 3.32VTGRR2625 pKa = 11.84LIEE2628 pKa = 4.21TVKK2631 pKa = 10.83LKK2633 pKa = 10.85SEE2635 pKa = 4.24EE2636 pKa = 4.03KK2637 pKa = 10.58QEE2639 pKa = 4.28VIDD2642 pKa = 3.81MSHH2645 pKa = 7.08WDD2647 pKa = 3.05SAMYY2651 pKa = 10.61LFVIKK2656 pKa = 10.44SDD2658 pKa = 3.49LGQVTKK2664 pKa = 10.9SVVKK2668 pKa = 10.13EE2669 pKa = 3.84
Molecular weight: 274.11 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5VWL3|A0A0C5VWL3_9FLAO Membrane protein OS=Siansivirga zeaxanthinifaciens CC-SAMT-1 OX=1454006 GN=AW14_07575 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.6LSVSSEE48 pKa = 4.04SRR50 pKa = 11.84HH51 pKa = 5.63KK52 pKa = 10.69KK53 pKa = 9.55
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.6LSVSSEE48 pKa = 4.04SRR50 pKa = 11.84HH51 pKa = 5.63KK52 pKa = 10.69KK53 pKa = 9.55
Molecular weight: 6.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
966228 |
49 |
4610 |
352.1 |
39.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.409 ± 0.049 | 0.77 ± 0.017 |
5.583 ± 0.038 | 6.275 ± 0.052 |
5.257 ± 0.039 | 6.323 ± 0.046 |
1.75 ± 0.023 | 8.132 ± 0.045 |
7.891 ± 0.07 | 9.228 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.039 ± 0.025 | 6.723 ± 0.05 |
3.378 ± 0.026 | 3.269 ± 0.022 |
3.16 ± 0.029 | 6.462 ± 0.037 |
6.033 ± 0.065 | 6.213 ± 0.043 |
1.048 ± 0.016 | 4.059 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
