
Vibrio phage 2E1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
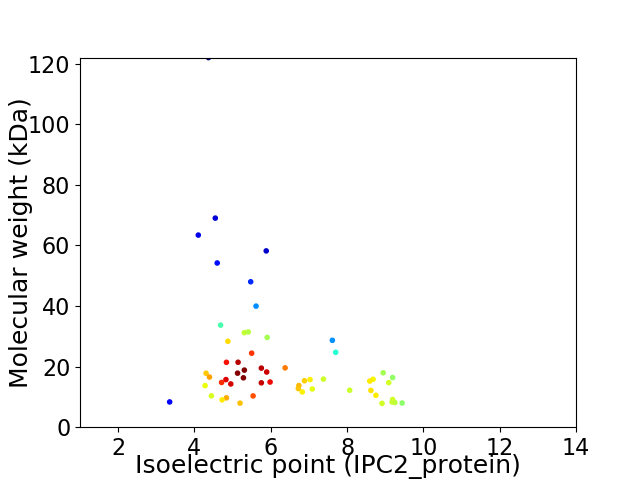
Virtual 2D-PAGE plot for 57 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
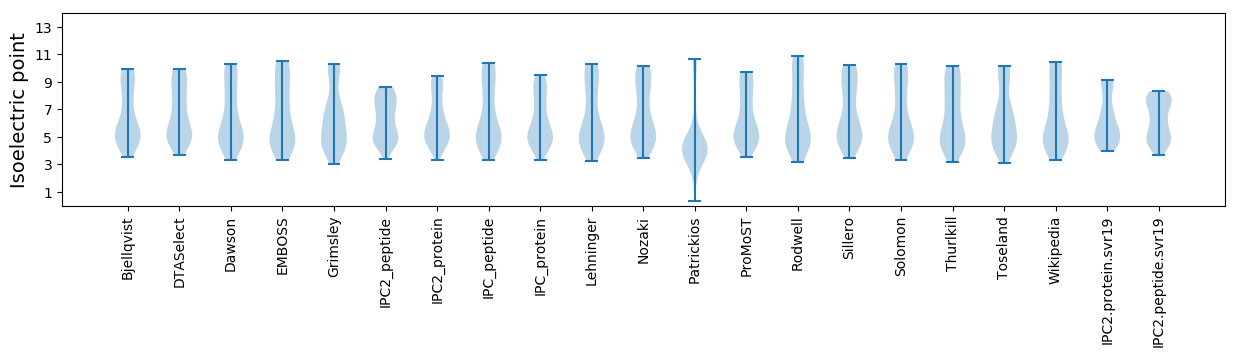
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1C9LW00|A0A1C9LW00_9CAUD Uncharacterized protein OS=Vibrio phage 2E1 OX=1873904 PE=4 SV=1
MM1 pKa = 6.8TLKK4 pKa = 10.71VVDD7 pKa = 5.31PINITEE13 pKa = 4.31AVLTASDD20 pKa = 3.9IPEE23 pKa = 4.61PDD25 pKa = 3.13ASVGEE30 pKa = 4.58VEE32 pKa = 4.24WQDD35 pKa = 3.29PSVLGRR41 pKa = 11.84FGNLPDD47 pKa = 3.48VQYY50 pKa = 10.46TVVLGSDD57 pKa = 3.6GFLYY61 pKa = 10.94SFGRR65 pKa = 11.84AGAVVKK71 pKa = 10.34IDD73 pKa = 4.2PDD75 pKa = 3.79TSTMTTFGSYY85 pKa = 10.19NVGRR89 pKa = 11.84VYY91 pKa = 10.97AAVLAGDD98 pKa = 3.81GNIYY102 pKa = 10.81AVGSGSAVLKK112 pKa = 10.9LEE114 pKa = 4.48ISTQTLSTFGSYY126 pKa = 10.78AGTCEE131 pKa = 4.17SATLAPDD138 pKa = 3.46GDD140 pKa = 4.34IYY142 pKa = 11.33CVSTNGFVLKK152 pKa = 10.08IDD154 pKa = 3.93VSAQTTSQFGSYY166 pKa = 9.59GAGYY170 pKa = 7.83KK171 pKa = 9.47TSSLGGDD178 pKa = 3.51GNIYY182 pKa = 10.39CAGSQGDD189 pKa = 3.84VLKK192 pKa = 10.51IDD194 pKa = 3.61VTEE197 pKa = 4.13QTVSTFGSVTGKK209 pKa = 10.9YY210 pKa = 9.19YY211 pKa = 11.17GSAQGVDD218 pKa = 3.26GNVYY222 pKa = 10.61CIGYY226 pKa = 9.18SGNVLKK232 pKa = 10.9INVSNQSVSTFGSCVGAFEE251 pKa = 5.27SAALDD256 pKa = 3.37ANGRR260 pKa = 11.84IKK262 pKa = 10.64CVASSSTTASGDD274 pKa = 3.12ILEE277 pKa = 5.15IDD279 pKa = 3.7TASEE283 pKa = 3.88SVYY286 pKa = 10.76RR287 pKa = 11.84FGSYY291 pKa = 9.32TDD293 pKa = 3.58NFFSIALAPNGKK305 pKa = 8.45MFAVGNDD312 pKa = 3.37VGSSTGIIEE321 pKa = 4.57ILDD324 pKa = 3.85TYY326 pKa = 11.52SPGDD330 pKa = 3.42RR331 pKa = 11.84VALEE335 pKa = 4.35SEE337 pKa = 4.13HH338 pKa = 7.38LVYY341 pKa = 10.66QCLSVTYY348 pKa = 9.34QNPKK352 pKa = 9.96VGATEE357 pKa = 5.07DD358 pKa = 3.81STATWIVVGPTNKK371 pKa = 8.94WAMFDD376 pKa = 3.56GLQNTKK382 pKa = 9.54TSSSTDD388 pKa = 3.5FTITLNPITYY398 pKa = 9.99VNTLAMFGFSGVEE411 pKa = 3.89SVRR414 pKa = 11.84IEE416 pKa = 3.89VDD418 pKa = 2.61NSLDD422 pKa = 3.39VNIYY426 pKa = 10.7DD427 pKa = 3.92KK428 pKa = 10.39TYY430 pKa = 11.17SMSDD434 pKa = 3.1FSAIYY439 pKa = 10.32DD440 pKa = 3.39HH441 pKa = 6.09YY442 pKa = 10.06TYY444 pKa = 11.22VFYY447 pKa = 10.88QIASLDD453 pKa = 3.82DD454 pKa = 5.39FIATDD459 pKa = 5.33LPPLPNTTIRR469 pKa = 11.84VTFSGSNMTIGEE481 pKa = 4.67LVTGFAIDD489 pKa = 3.28IGQLVAEE496 pKa = 4.52STKK499 pKa = 10.54SDD501 pKa = 3.04RR502 pKa = 11.84FRR504 pKa = 11.84YY505 pKa = 9.58RR506 pKa = 11.84EE507 pKa = 3.62QEE509 pKa = 4.14YY510 pKa = 11.23NEE512 pKa = 4.02FGYY515 pKa = 8.4PTGEE519 pKa = 3.75APIVVEE525 pKa = 4.1LNTYY529 pKa = 10.43DD530 pKa = 3.81VLVPKK535 pKa = 10.58LNNQAIQKK543 pKa = 10.39LLDD546 pKa = 3.59TLTGEE551 pKa = 3.95NTLWVGDD558 pKa = 3.63IGGGQSLVTYY568 pKa = 10.19GLFEE572 pKa = 4.75RR573 pKa = 11.84SPIPYY578 pKa = 10.38SMPNHH583 pKa = 6.54INYY586 pKa = 9.94QITVRR591 pKa = 11.84ASVV594 pKa = 2.9
MM1 pKa = 6.8TLKK4 pKa = 10.71VVDD7 pKa = 5.31PINITEE13 pKa = 4.31AVLTASDD20 pKa = 3.9IPEE23 pKa = 4.61PDD25 pKa = 3.13ASVGEE30 pKa = 4.58VEE32 pKa = 4.24WQDD35 pKa = 3.29PSVLGRR41 pKa = 11.84FGNLPDD47 pKa = 3.48VQYY50 pKa = 10.46TVVLGSDD57 pKa = 3.6GFLYY61 pKa = 10.94SFGRR65 pKa = 11.84AGAVVKK71 pKa = 10.34IDD73 pKa = 4.2PDD75 pKa = 3.79TSTMTTFGSYY85 pKa = 10.19NVGRR89 pKa = 11.84VYY91 pKa = 10.97AAVLAGDD98 pKa = 3.81GNIYY102 pKa = 10.81AVGSGSAVLKK112 pKa = 10.9LEE114 pKa = 4.48ISTQTLSTFGSYY126 pKa = 10.78AGTCEE131 pKa = 4.17SATLAPDD138 pKa = 3.46GDD140 pKa = 4.34IYY142 pKa = 11.33CVSTNGFVLKK152 pKa = 10.08IDD154 pKa = 3.93VSAQTTSQFGSYY166 pKa = 9.59GAGYY170 pKa = 7.83KK171 pKa = 9.47TSSLGGDD178 pKa = 3.51GNIYY182 pKa = 10.39CAGSQGDD189 pKa = 3.84VLKK192 pKa = 10.51IDD194 pKa = 3.61VTEE197 pKa = 4.13QTVSTFGSVTGKK209 pKa = 10.9YY210 pKa = 9.19YY211 pKa = 11.17GSAQGVDD218 pKa = 3.26GNVYY222 pKa = 10.61CIGYY226 pKa = 9.18SGNVLKK232 pKa = 10.9INVSNQSVSTFGSCVGAFEE251 pKa = 5.27SAALDD256 pKa = 3.37ANGRR260 pKa = 11.84IKK262 pKa = 10.64CVASSSTTASGDD274 pKa = 3.12ILEE277 pKa = 5.15IDD279 pKa = 3.7TASEE283 pKa = 3.88SVYY286 pKa = 10.76RR287 pKa = 11.84FGSYY291 pKa = 9.32TDD293 pKa = 3.58NFFSIALAPNGKK305 pKa = 8.45MFAVGNDD312 pKa = 3.37VGSSTGIIEE321 pKa = 4.57ILDD324 pKa = 3.85TYY326 pKa = 11.52SPGDD330 pKa = 3.42RR331 pKa = 11.84VALEE335 pKa = 4.35SEE337 pKa = 4.13HH338 pKa = 7.38LVYY341 pKa = 10.66QCLSVTYY348 pKa = 9.34QNPKK352 pKa = 9.96VGATEE357 pKa = 5.07DD358 pKa = 3.81STATWIVVGPTNKK371 pKa = 8.94WAMFDD376 pKa = 3.56GLQNTKK382 pKa = 9.54TSSSTDD388 pKa = 3.5FTITLNPITYY398 pKa = 9.99VNTLAMFGFSGVEE411 pKa = 3.89SVRR414 pKa = 11.84IEE416 pKa = 3.89VDD418 pKa = 2.61NSLDD422 pKa = 3.39VNIYY426 pKa = 10.7DD427 pKa = 3.92KK428 pKa = 10.39TYY430 pKa = 11.17SMSDD434 pKa = 3.1FSAIYY439 pKa = 10.32DD440 pKa = 3.39HH441 pKa = 6.09YY442 pKa = 10.06TYY444 pKa = 11.22VFYY447 pKa = 10.88QIASLDD453 pKa = 3.82DD454 pKa = 5.39FIATDD459 pKa = 5.33LPPLPNTTIRR469 pKa = 11.84VTFSGSNMTIGEE481 pKa = 4.67LVTGFAIDD489 pKa = 3.28IGQLVAEE496 pKa = 4.52STKK499 pKa = 10.54SDD501 pKa = 3.04RR502 pKa = 11.84FRR504 pKa = 11.84YY505 pKa = 9.58RR506 pKa = 11.84EE507 pKa = 3.62QEE509 pKa = 4.14YY510 pKa = 11.23NEE512 pKa = 4.02FGYY515 pKa = 8.4PTGEE519 pKa = 3.75APIVVEE525 pKa = 4.1LNTYY529 pKa = 10.43DD530 pKa = 3.81VLVPKK535 pKa = 10.58LNNQAIQKK543 pKa = 10.39LLDD546 pKa = 3.59TLTGEE551 pKa = 3.95NTLWVGDD558 pKa = 3.63IGGGQSLVTYY568 pKa = 10.19GLFEE572 pKa = 4.75RR573 pKa = 11.84SPIPYY578 pKa = 10.38SMPNHH583 pKa = 6.54INYY586 pKa = 9.94QITVRR591 pKa = 11.84ASVV594 pKa = 2.9
Molecular weight: 63.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1C9LVU1|A0A1C9LVU1_9CAUD Uncharacterized protein OS=Vibrio phage 2E1 OX=1873904 PE=4 SV=1
MM1 pKa = 7.45WLISFKK7 pKa = 10.29GLRR10 pKa = 11.84MNNVFRR16 pKa = 11.84MPKK19 pKa = 9.49LHH21 pKa = 6.82RR22 pKa = 11.84NSAAMGKK29 pKa = 9.15ILKK32 pKa = 10.13DD33 pKa = 3.1IEE35 pKa = 4.17KK36 pKa = 10.34RR37 pKa = 11.84GCFYY41 pKa = 11.35YY42 pKa = 10.84NNQRR46 pKa = 11.84VKK48 pKa = 10.83LDD50 pKa = 3.52EE51 pKa = 5.34ANTKK55 pKa = 9.7VDD57 pKa = 3.2GVYY60 pKa = 10.67VYY62 pKa = 10.77LAYY65 pKa = 9.68TPAA68 pKa = 5.34
MM1 pKa = 7.45WLISFKK7 pKa = 10.29GLRR10 pKa = 11.84MNNVFRR16 pKa = 11.84MPKK19 pKa = 9.49LHH21 pKa = 6.82RR22 pKa = 11.84NSAAMGKK29 pKa = 9.15ILKK32 pKa = 10.13DD33 pKa = 3.1IEE35 pKa = 4.17KK36 pKa = 10.34RR37 pKa = 11.84GCFYY41 pKa = 11.35YY42 pKa = 10.84NNQRR46 pKa = 11.84VKK48 pKa = 10.83LDD50 pKa = 3.52EE51 pKa = 5.34ANTKK55 pKa = 9.7VDD57 pKa = 3.2GVYY60 pKa = 10.67VYY62 pKa = 10.77LAYY65 pKa = 9.68TPAA68 pKa = 5.34
Molecular weight: 7.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11377 |
68 |
1142 |
199.6 |
22.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.612 ± 0.628 | 1.406 ± 0.217 |
6.311 ± 0.208 | 6.777 ± 0.341 |
3.964 ± 0.231 | 6.909 ± 0.356 |
1.828 ± 0.252 | 6.267 ± 0.209 |
6.803 ± 0.414 | 7.682 ± 0.292 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.971 ± 0.202 | 5.344 ± 0.171 |
3.068 ± 0.257 | 3.911 ± 0.366 |
4.158 ± 0.288 | 7.304 ± 0.343 |
6.056 ± 0.328 | 6.452 ± 0.281 |
1.292 ± 0.146 | 3.885 ± 0.233 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
