
Apis mellifera associated microvirus 50
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.79
Get precalculated fractions of proteins
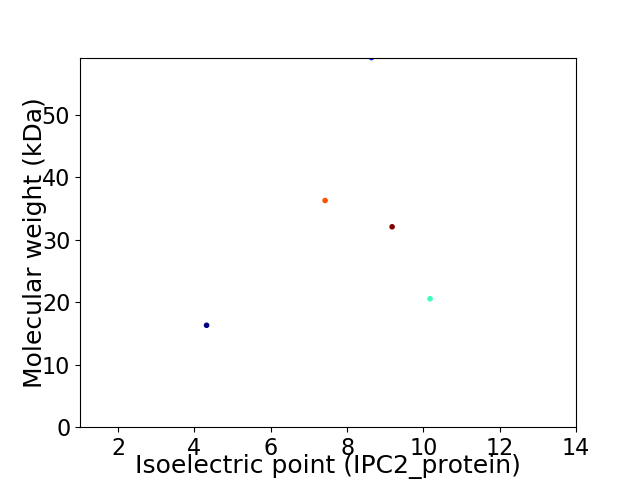
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S8UU36|A0A3S8UU36_9VIRU Major capsid protein OS=Apis mellifera associated microvirus 50 OX=2494781 PE=3 SV=1
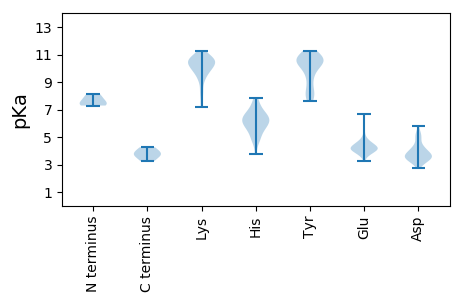
MM1 pKa = 7.29EE2 pKa = 6.15LSMKK6 pKa = 10.37KK7 pKa = 10.14LSEE10 pKa = 4.12VVKK13 pKa = 10.54EE14 pKa = 4.07FFGKK18 pKa = 10.17SRR20 pKa = 11.84LDD22 pKa = 3.13EE23 pKa = 4.68RR24 pKa = 11.84YY25 pKa = 9.55PGGSPLEE32 pKa = 4.2VCNPQPVEE40 pKa = 3.97VPLRR44 pKa = 11.84FQRR47 pKa = 11.84PPTMEE52 pKa = 3.27EE53 pKa = 4.05RR54 pKa = 11.84IRR56 pKa = 11.84QFIRR60 pKa = 11.84SEE62 pKa = 3.73AMRR65 pKa = 11.84RR66 pKa = 11.84EE67 pKa = 4.15AEE69 pKa = 3.98AQGAEE74 pKa = 4.27TFEE77 pKa = 4.16EE78 pKa = 4.56ADD80 pKa = 4.06DD81 pKa = 5.08FEE83 pKa = 6.68VDD85 pKa = 5.39DD86 pKa = 5.35PDD88 pKa = 4.82FDD90 pKa = 4.23VPSSEE95 pKa = 4.28FEE97 pKa = 4.23VMEE100 pKa = 5.4LEE102 pKa = 4.48PQDD105 pKa = 4.41RR106 pKa = 11.84DD107 pKa = 3.3AAPPAEE113 pKa = 4.52PTSPQPPAAAPGPAQGAGPSGTPPAPAGGVPAAPGPAASS152 pKa = 3.94
MM1 pKa = 7.29EE2 pKa = 6.15LSMKK6 pKa = 10.37KK7 pKa = 10.14LSEE10 pKa = 4.12VVKK13 pKa = 10.54EE14 pKa = 4.07FFGKK18 pKa = 10.17SRR20 pKa = 11.84LDD22 pKa = 3.13EE23 pKa = 4.68RR24 pKa = 11.84YY25 pKa = 9.55PGGSPLEE32 pKa = 4.2VCNPQPVEE40 pKa = 3.97VPLRR44 pKa = 11.84FQRR47 pKa = 11.84PPTMEE52 pKa = 3.27EE53 pKa = 4.05RR54 pKa = 11.84IRR56 pKa = 11.84QFIRR60 pKa = 11.84SEE62 pKa = 3.73AMRR65 pKa = 11.84RR66 pKa = 11.84EE67 pKa = 4.15AEE69 pKa = 3.98AQGAEE74 pKa = 4.27TFEE77 pKa = 4.16EE78 pKa = 4.56ADD80 pKa = 4.06DD81 pKa = 5.08FEE83 pKa = 6.68VDD85 pKa = 5.39DD86 pKa = 5.35PDD88 pKa = 4.82FDD90 pKa = 4.23VPSSEE95 pKa = 4.28FEE97 pKa = 4.23VMEE100 pKa = 5.4LEE102 pKa = 4.48PQDD105 pKa = 4.41RR106 pKa = 11.84DD107 pKa = 3.3AAPPAEE113 pKa = 4.52PTSPQPPAAAPGPAQGAGPSGTPPAPAGGVPAAPGPAASS152 pKa = 3.94
Molecular weight: 16.3 kDa
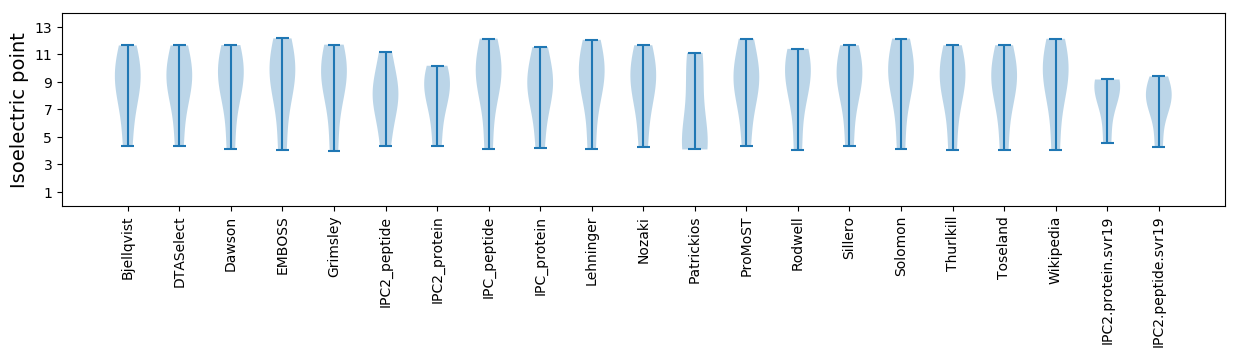
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S8UU72|A0A3S8UU72_9VIRU DNA pilot protein OS=Apis mellifera associated microvirus 50 OX=2494781 PE=4 SV=1
HHH2 pKa = 8.13IPHHH6 pKa = 4.78KKK8 pKa = 9.71RR9 pKa = 11.84YYY11 pKa = 7.96LKKK14 pKa = 10.17GVMEEE19 pKa = 5.03GCGRR23 pKa = 11.84CMPCRR28 pKa = 11.84INRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84EEE35 pKa = 3.6TSRR38 pKa = 11.84MMLEEE43 pKa = 4.3QGHHH47 pKa = 4.76VSWFVTLTYYY57 pKa = 10.65PEEE60 pKa = 3.98LPEEE64 pKa = 4.51GTLVPKKK71 pKa = 10.34HH72 pKa = 5.61QDDD75 pKa = 2.73LKKK78 pKa = 10.7LRR80 pKa = 11.84TATGEEE86 pKa = 4.03KK87 pKa = 9.64RR88 pKa = 11.84FYYY91 pKa = 11.25VGEEE95 pKa = 4.39YY96 pKa = 10.64DDD98 pKa = 4.02KK99 pKa = 10.01WRR101 pKa = 11.84PHHH104 pKa = 3.78YY105 pKa = 10.2HH106 pKa = 6.94ILFGGPSTPDDD117 pKa = 4.21ILATWHHH124 pKa = 6.53GLGHHH129 pKa = 5.98GFVTAEEE136 pKa = 4.06IQYYY140 pKa = 7.97ASYYY144 pKa = 10.33EEE146 pKa = 4.61KK147 pKa = 10.91KK148 pKa = 7.21TSSKKK153 pKa = 10.87DD154 pKa = 3.09DD155 pKa = 3.85LNGRR159 pKa = 11.84YYY161 pKa = 9.31EEE163 pKa = 4.18ARR165 pKa = 11.84MSRR168 pKa = 11.84KKK170 pKa = 9.59GIGALAVDDD179 pKa = 3.86MFGKKK184 pKa = 10.68LTSKKK189 pKa = 10.54GVDDD193 pKa = 3.6IIRR196 pKa = 11.84EEE198 pKa = 4.09DDD200 pKa = 3.61PSMVRR205 pKa = 11.84FEEE208 pKa = 4.28KKK210 pKa = 9.11KK211 pKa = 8.96PLGRR215 pKa = 11.84YYY217 pKa = 8.2RR218 pKa = 11.84SRR220 pKa = 11.84ARR222 pKa = 11.84AAAGFEEE229 pKa = 4.09EE230 pKa = 5.54GMPEEE235 pKa = 4.57DD236 pKa = 5.03FIQMALEEE244 pKa = 4.17QATLSDDD251 pKa = 3.35EEE253 pKa = 3.75FYYY256 pKa = 10.7HHH258 pKa = 5.78EE259 pKa = 4.47DDD261 pKa = 4.48VNQEEE266 pKa = 3.3TAKKK270 pKa = 10.25RR271 pKa = 11.84AKKK274 pKa = 9.92RR275 pKa = 11.84ASKKK279 pKa = 10.61SI
HHH2 pKa = 8.13IPHHH6 pKa = 4.78KKK8 pKa = 9.71RR9 pKa = 11.84YYY11 pKa = 7.96LKKK14 pKa = 10.17GVMEEE19 pKa = 5.03GCGRR23 pKa = 11.84CMPCRR28 pKa = 11.84INRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84EEE35 pKa = 3.6TSRR38 pKa = 11.84MMLEEE43 pKa = 4.3QGHHH47 pKa = 4.76VSWFVTLTYYY57 pKa = 10.65PEEE60 pKa = 3.98LPEEE64 pKa = 4.51GTLVPKKK71 pKa = 10.34HH72 pKa = 5.61QDDD75 pKa = 2.73LKKK78 pKa = 10.7LRR80 pKa = 11.84TATGEEE86 pKa = 4.03KK87 pKa = 9.64RR88 pKa = 11.84FYYY91 pKa = 11.25VGEEE95 pKa = 4.39YY96 pKa = 10.64DDD98 pKa = 4.02KK99 pKa = 10.01WRR101 pKa = 11.84PHHH104 pKa = 3.78YY105 pKa = 10.2HH106 pKa = 6.94ILFGGPSTPDDD117 pKa = 4.21ILATWHHH124 pKa = 6.53GLGHHH129 pKa = 5.98GFVTAEEE136 pKa = 4.06IQYYY140 pKa = 7.97ASYYY144 pKa = 10.33EEE146 pKa = 4.61KK147 pKa = 10.91KK148 pKa = 7.21TSSKKK153 pKa = 10.87DD154 pKa = 3.09DD155 pKa = 3.85LNGRR159 pKa = 11.84YYY161 pKa = 9.31EEE163 pKa = 4.18ARR165 pKa = 11.84MSRR168 pKa = 11.84KKK170 pKa = 9.59GIGALAVDDD179 pKa = 3.86MFGKKK184 pKa = 10.68LTSKKK189 pKa = 10.54GVDDD193 pKa = 3.6IIRR196 pKa = 11.84EEE198 pKa = 4.09DDD200 pKa = 3.61PSMVRR205 pKa = 11.84FEEE208 pKa = 4.28KKK210 pKa = 9.11KK211 pKa = 8.96PLGRR215 pKa = 11.84YYY217 pKa = 8.2RR218 pKa = 11.84SRR220 pKa = 11.84ARR222 pKa = 11.84AAAGFEEE229 pKa = 4.09EE230 pKa = 5.54GMPEEE235 pKa = 4.57DD236 pKa = 5.03FIQMALEEE244 pKa = 4.17QATLSDDD251 pKa = 3.35EEE253 pKa = 3.75FYYY256 pKa = 10.7HHH258 pKa = 5.78EE259 pKa = 4.47DDD261 pKa = 4.48VNQEEE266 pKa = 3.3TAKKK270 pKa = 10.25RR271 pKa = 11.84AKKK274 pKa = 9.92RR275 pKa = 11.84ASKKK279 pKa = 10.61SI
Molecular weight: 32.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1480 |
152 |
525 |
296.0 |
32.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.932 ± 1.664 | 0.811 ± 0.237 |
4.73 ± 0.382 | 6.014 ± 1.209 |
3.311 ± 0.528 | 8.108 ± 0.532 |
2.432 ± 0.404 | 3.581 ± 0.723 |
3.784 ± 0.58 | 7.365 ± 0.561 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.568 ± 0.297 | 2.5 ± 0.529 |
7.365 ± 1.58 | 3.243 ± 0.582 |
9.459 ± 1.734 | 8.378 ± 0.886 |
4.459 ± 0.305 | 6.689 ± 0.33 |
1.824 ± 0.344 | 3.446 ± 0.734 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
