
Arenibacter nanhaiticus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Arenibacter
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
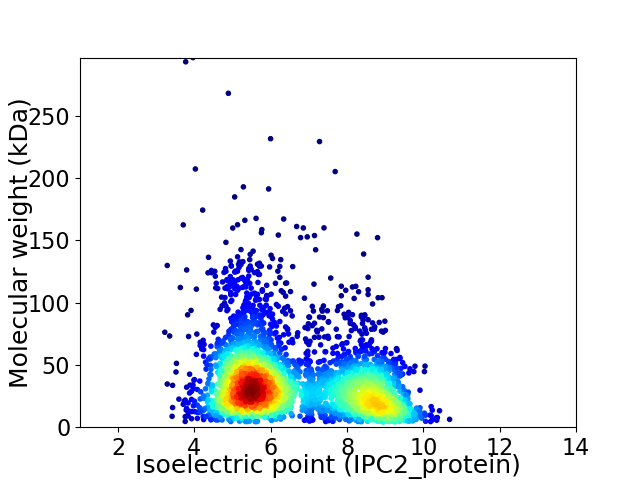
Virtual 2D-PAGE plot for 3553 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M6I4C6|A0A1M6I4C6_9FLAO Glycerol kinase OS=Arenibacter nanhaiticus OX=558155 GN=glpK PE=3 SV=1
MM1 pKa = 7.18EE2 pKa = 5.06HH3 pKa = 6.91SISIFAFKK11 pKa = 10.44KK12 pKa = 9.57SSCSGTLGKK21 pKa = 10.68LFFLGFFLLTSLSFAQQADD40 pKa = 3.79LRR42 pKa = 11.84SHH44 pKa = 4.52VTEE47 pKa = 4.79GQDD50 pKa = 3.28CTSNDD55 pKa = 3.21VQLISAQLSSEE66 pKa = 4.05EE67 pKa = 4.55CTTCSPGDD75 pKa = 3.64ILVVDD80 pKa = 4.26LLITVRR86 pKa = 11.84HH87 pKa = 5.12NTNSVRR93 pKa = 11.84PSLAVFGDD101 pKa = 4.24LTTTLPDD108 pKa = 3.78GSSTTSVFTEE118 pKa = 4.22CGGPIIPKK126 pKa = 10.31DD127 pKa = 3.41KK128 pKa = 10.41VLNGIPVDD136 pKa = 3.4KK137 pKa = 10.43GGNGIQTIKK146 pKa = 10.22YY147 pKa = 7.94GQVSYY152 pKa = 10.95ICGSKK157 pKa = 11.03LEE159 pKa = 4.02VDD161 pKa = 4.45NILVAWTVPPDD172 pKa = 4.12GGCPIADD179 pKa = 3.89ASPHH183 pKa = 5.99PKK185 pKa = 10.21CGFQEE190 pKa = 4.52GSLIIIPPLHH200 pKa = 6.91AIAEE204 pKa = 4.41AFCTPEE210 pKa = 3.78NYY212 pKa = 9.38IDD214 pKa = 4.19LEE216 pKa = 4.72AIGGSAPYY224 pKa = 9.69TYY226 pKa = 10.54DD227 pKa = 2.71WKK229 pKa = 11.13GPNGFTSTDD238 pKa = 3.08EE239 pKa = 4.44DD240 pKa = 4.51LFGVLPGNYY249 pKa = 9.29DD250 pKa = 3.46VTVTDD255 pKa = 5.25SKK257 pKa = 11.33GCSINTSVTKK267 pKa = 10.52IKK269 pKa = 10.5CCEE272 pKa = 3.94FFATPNLDD280 pKa = 3.17SSEE283 pKa = 4.03QNIEE287 pKa = 3.95GCDD290 pKa = 3.78VSDD293 pKa = 4.52LPSPFTDD300 pKa = 3.58PEE302 pKa = 4.31NVFTDD307 pKa = 3.28ITEE310 pKa = 4.33NPCGNLIMHH319 pKa = 6.68QSDD322 pKa = 3.38ILNGNLCTGGLIINRR337 pKa = 11.84TYY339 pKa = 11.2TLFDD343 pKa = 4.46DD344 pKa = 5.12LNNNNTLDD352 pKa = 3.68SGEE355 pKa = 4.23EE356 pKa = 3.93FAVSYY361 pKa = 10.7QNFRR365 pKa = 11.84IIDD368 pKa = 3.63TTLPTFNTPLPEE380 pKa = 5.17DD381 pKa = 3.42STAAFDD387 pKa = 5.21NIPSPPTLSANDD399 pKa = 3.55NCDD402 pKa = 3.79DD403 pKa = 4.03NVSVTLVEE411 pKa = 4.38TYY413 pKa = 10.37IGDD416 pKa = 4.07STSTTYY422 pKa = 10.61TIVRR426 pKa = 11.84TWTASDD432 pKa = 3.68CAGNEE437 pKa = 4.35TVHH440 pKa = 5.34TQYY443 pKa = 10.95IYY445 pKa = 9.79VTEE448 pKa = 4.34NGDD451 pKa = 4.23PIGLKK456 pKa = 10.35INDD459 pKa = 3.69VTVDD463 pKa = 3.47EE464 pKa = 5.08ADD466 pKa = 3.28NTAVFTVTLTGRR478 pKa = 11.84VSGGFNVDD486 pKa = 3.65YY487 pKa = 11.36NSVNQTAIAPNDD499 pKa = 3.4FTAIGITSLSFSGNHH514 pKa = 5.81GEE516 pKa = 4.3KK517 pKa = 9.17KK518 pKa = 9.17TISVSIKK525 pKa = 9.3EE526 pKa = 4.17DD527 pKa = 3.24QFVEE531 pKa = 4.08DD532 pKa = 5.52DD533 pKa = 3.55EE534 pKa = 4.98TFTIDD539 pKa = 5.04LSNLSTTEE547 pKa = 3.51IPINKK552 pKa = 8.49ATGTGTIIDD561 pKa = 4.32NDD563 pKa = 3.92SATISITDD571 pKa = 3.43MEE573 pKa = 4.84INEE576 pKa = 4.22GTGIFNIQVTLIGKK590 pKa = 6.38TQDD593 pKa = 2.98PFTINFSTADD603 pKa = 3.37GSAIKK608 pKa = 10.59VDD610 pKa = 4.58DD611 pKa = 3.82YY612 pKa = 12.01VEE614 pKa = 5.17AIGQITFPANSTNGTVQNISLEE636 pKa = 4.34VIDD639 pKa = 5.5DD640 pKa = 4.31NIIEE644 pKa = 4.26FTEE647 pKa = 4.01NFLVNLNSISGFGDD661 pKa = 2.87ISFLDD666 pKa = 3.53NQANIDD672 pKa = 4.31IIDD675 pKa = 3.76NDD677 pKa = 4.88AITGTGISFDD687 pKa = 3.63NTNVIVNEE695 pKa = 4.23DD696 pKa = 3.32AGTAIFTVRR705 pKa = 11.84LTGQVFGGFTLDD717 pKa = 3.54YY718 pKa = 8.29TTEE721 pKa = 4.11NVTAMAGSDD730 pKa = 3.47YY731 pKa = 10.31TAEE734 pKa = 4.03SGKK737 pKa = 10.94LNFAGNDD744 pKa = 3.64NEE746 pKa = 4.08FHH748 pKa = 6.45TIEE751 pKa = 4.52VPIIDD756 pKa = 4.45DD757 pKa = 3.69TLIEE761 pKa = 4.14PTEE764 pKa = 4.15TFVVNLSNIDD774 pKa = 3.64PAIVSINTAQARR786 pKa = 11.84GTIKK790 pKa = 11.15DD791 pKa = 3.48NDD793 pKa = 3.54ATAGTGVSFKK803 pKa = 10.4NTDD806 pKa = 2.61ITVNEE811 pKa = 4.38DD812 pKa = 2.66AGTATFKK819 pKa = 11.02VVLSGNVQGGFTLDD833 pKa = 3.46YY834 pKa = 8.43TTDD837 pKa = 3.31NGSAVQPDD845 pKa = 5.42DD846 pKa = 3.41YY847 pKa = 11.33TLTYY851 pKa = 9.08DD852 pKa = 3.25TLTFVGNDD860 pKa = 3.79GEE862 pKa = 4.75TKK864 pKa = 10.52DD865 pKa = 3.37ITVPIIDD872 pKa = 5.17DD873 pKa = 3.97LLIEE877 pKa = 4.11PTEE880 pKa = 3.96NFFVKK885 pKa = 10.59LSNLSTSLIGINTPQAKK902 pKa = 10.32GNIIDD907 pKa = 3.93NDD909 pKa = 4.38AIAGTGIAFTNTNVTVTEE927 pKa = 4.22GTDD930 pKa = 2.75AFAIFEE936 pKa = 4.33VTLTGDD942 pKa = 2.61ISEE945 pKa = 4.57NVSVDD950 pKa = 3.13YY951 pKa = 8.25TTIDD955 pKa = 3.3GTALDD960 pKa = 4.3PSDD963 pKa = 4.27YY964 pKa = 10.57LTTTSTITFTPTIKK978 pKa = 10.32SYY980 pKa = 10.36EE981 pKa = 4.0IQVPIMDD988 pKa = 4.72DD989 pKa = 3.97AIIEE993 pKa = 4.13PSEE996 pKa = 4.08AFTVEE1001 pKa = 4.12LSNIQSNLGIGFVDD1015 pKa = 4.74GNATNTATGNILDD1028 pKa = 4.78DD1029 pKa = 4.54DD1030 pKa = 4.55AVGGTGIAFTYY1041 pKa = 10.23TNVTVTEE1048 pKa = 4.24GTDD1051 pKa = 2.99AFAA1054 pKa = 6.04
MM1 pKa = 7.18EE2 pKa = 5.06HH3 pKa = 6.91SISIFAFKK11 pKa = 10.44KK12 pKa = 9.57SSCSGTLGKK21 pKa = 10.68LFFLGFFLLTSLSFAQQADD40 pKa = 3.79LRR42 pKa = 11.84SHH44 pKa = 4.52VTEE47 pKa = 4.79GQDD50 pKa = 3.28CTSNDD55 pKa = 3.21VQLISAQLSSEE66 pKa = 4.05EE67 pKa = 4.55CTTCSPGDD75 pKa = 3.64ILVVDD80 pKa = 4.26LLITVRR86 pKa = 11.84HH87 pKa = 5.12NTNSVRR93 pKa = 11.84PSLAVFGDD101 pKa = 4.24LTTTLPDD108 pKa = 3.78GSSTTSVFTEE118 pKa = 4.22CGGPIIPKK126 pKa = 10.31DD127 pKa = 3.41KK128 pKa = 10.41VLNGIPVDD136 pKa = 3.4KK137 pKa = 10.43GGNGIQTIKK146 pKa = 10.22YY147 pKa = 7.94GQVSYY152 pKa = 10.95ICGSKK157 pKa = 11.03LEE159 pKa = 4.02VDD161 pKa = 4.45NILVAWTVPPDD172 pKa = 4.12GGCPIADD179 pKa = 3.89ASPHH183 pKa = 5.99PKK185 pKa = 10.21CGFQEE190 pKa = 4.52GSLIIIPPLHH200 pKa = 6.91AIAEE204 pKa = 4.41AFCTPEE210 pKa = 3.78NYY212 pKa = 9.38IDD214 pKa = 4.19LEE216 pKa = 4.72AIGGSAPYY224 pKa = 9.69TYY226 pKa = 10.54DD227 pKa = 2.71WKK229 pKa = 11.13GPNGFTSTDD238 pKa = 3.08EE239 pKa = 4.44DD240 pKa = 4.51LFGVLPGNYY249 pKa = 9.29DD250 pKa = 3.46VTVTDD255 pKa = 5.25SKK257 pKa = 11.33GCSINTSVTKK267 pKa = 10.52IKK269 pKa = 10.5CCEE272 pKa = 3.94FFATPNLDD280 pKa = 3.17SSEE283 pKa = 4.03QNIEE287 pKa = 3.95GCDD290 pKa = 3.78VSDD293 pKa = 4.52LPSPFTDD300 pKa = 3.58PEE302 pKa = 4.31NVFTDD307 pKa = 3.28ITEE310 pKa = 4.33NPCGNLIMHH319 pKa = 6.68QSDD322 pKa = 3.38ILNGNLCTGGLIINRR337 pKa = 11.84TYY339 pKa = 11.2TLFDD343 pKa = 4.46DD344 pKa = 5.12LNNNNTLDD352 pKa = 3.68SGEE355 pKa = 4.23EE356 pKa = 3.93FAVSYY361 pKa = 10.7QNFRR365 pKa = 11.84IIDD368 pKa = 3.63TTLPTFNTPLPEE380 pKa = 5.17DD381 pKa = 3.42STAAFDD387 pKa = 5.21NIPSPPTLSANDD399 pKa = 3.55NCDD402 pKa = 3.79DD403 pKa = 4.03NVSVTLVEE411 pKa = 4.38TYY413 pKa = 10.37IGDD416 pKa = 4.07STSTTYY422 pKa = 10.61TIVRR426 pKa = 11.84TWTASDD432 pKa = 3.68CAGNEE437 pKa = 4.35TVHH440 pKa = 5.34TQYY443 pKa = 10.95IYY445 pKa = 9.79VTEE448 pKa = 4.34NGDD451 pKa = 4.23PIGLKK456 pKa = 10.35INDD459 pKa = 3.69VTVDD463 pKa = 3.47EE464 pKa = 5.08ADD466 pKa = 3.28NTAVFTVTLTGRR478 pKa = 11.84VSGGFNVDD486 pKa = 3.65YY487 pKa = 11.36NSVNQTAIAPNDD499 pKa = 3.4FTAIGITSLSFSGNHH514 pKa = 5.81GEE516 pKa = 4.3KK517 pKa = 9.17KK518 pKa = 9.17TISVSIKK525 pKa = 9.3EE526 pKa = 4.17DD527 pKa = 3.24QFVEE531 pKa = 4.08DD532 pKa = 5.52DD533 pKa = 3.55EE534 pKa = 4.98TFTIDD539 pKa = 5.04LSNLSTTEE547 pKa = 3.51IPINKK552 pKa = 8.49ATGTGTIIDD561 pKa = 4.32NDD563 pKa = 3.92SATISITDD571 pKa = 3.43MEE573 pKa = 4.84INEE576 pKa = 4.22GTGIFNIQVTLIGKK590 pKa = 6.38TQDD593 pKa = 2.98PFTINFSTADD603 pKa = 3.37GSAIKK608 pKa = 10.59VDD610 pKa = 4.58DD611 pKa = 3.82YY612 pKa = 12.01VEE614 pKa = 5.17AIGQITFPANSTNGTVQNISLEE636 pKa = 4.34VIDD639 pKa = 5.5DD640 pKa = 4.31NIIEE644 pKa = 4.26FTEE647 pKa = 4.01NFLVNLNSISGFGDD661 pKa = 2.87ISFLDD666 pKa = 3.53NQANIDD672 pKa = 4.31IIDD675 pKa = 3.76NDD677 pKa = 4.88AITGTGISFDD687 pKa = 3.63NTNVIVNEE695 pKa = 4.23DD696 pKa = 3.32AGTAIFTVRR705 pKa = 11.84LTGQVFGGFTLDD717 pKa = 3.54YY718 pKa = 8.29TTEE721 pKa = 4.11NVTAMAGSDD730 pKa = 3.47YY731 pKa = 10.31TAEE734 pKa = 4.03SGKK737 pKa = 10.94LNFAGNDD744 pKa = 3.64NEE746 pKa = 4.08FHH748 pKa = 6.45TIEE751 pKa = 4.52VPIIDD756 pKa = 4.45DD757 pKa = 3.69TLIEE761 pKa = 4.14PTEE764 pKa = 4.15TFVVNLSNIDD774 pKa = 3.64PAIVSINTAQARR786 pKa = 11.84GTIKK790 pKa = 11.15DD791 pKa = 3.48NDD793 pKa = 3.54ATAGTGVSFKK803 pKa = 10.4NTDD806 pKa = 2.61ITVNEE811 pKa = 4.38DD812 pKa = 2.66AGTATFKK819 pKa = 11.02VVLSGNVQGGFTLDD833 pKa = 3.46YY834 pKa = 8.43TTDD837 pKa = 3.31NGSAVQPDD845 pKa = 5.42DD846 pKa = 3.41YY847 pKa = 11.33TLTYY851 pKa = 9.08DD852 pKa = 3.25TLTFVGNDD860 pKa = 3.79GEE862 pKa = 4.75TKK864 pKa = 10.52DD865 pKa = 3.37ITVPIIDD872 pKa = 5.17DD873 pKa = 3.97LLIEE877 pKa = 4.11PTEE880 pKa = 3.96NFFVKK885 pKa = 10.59LSNLSTSLIGINTPQAKK902 pKa = 10.32GNIIDD907 pKa = 3.93NDD909 pKa = 4.38AIAGTGIAFTNTNVTVTEE927 pKa = 4.22GTDD930 pKa = 2.75AFAIFEE936 pKa = 4.33VTLTGDD942 pKa = 2.61ISEE945 pKa = 4.57NVSVDD950 pKa = 3.13YY951 pKa = 8.25TTIDD955 pKa = 3.3GTALDD960 pKa = 4.3PSDD963 pKa = 4.27YY964 pKa = 10.57LTTTSTITFTPTIKK978 pKa = 10.32SYY980 pKa = 10.36EE981 pKa = 4.0IQVPIMDD988 pKa = 4.72DD989 pKa = 3.97AIIEE993 pKa = 4.13PSEE996 pKa = 4.08AFTVEE1001 pKa = 4.12LSNIQSNLGIGFVDD1015 pKa = 4.74GNATNTATGNILDD1028 pKa = 4.78DD1029 pKa = 4.54DD1030 pKa = 4.55AVGGTGIAFTYY1041 pKa = 10.23TNVTVTEE1048 pKa = 4.24GTDD1051 pKa = 2.99AFAA1054 pKa = 6.04
Molecular weight: 112.19 kDa
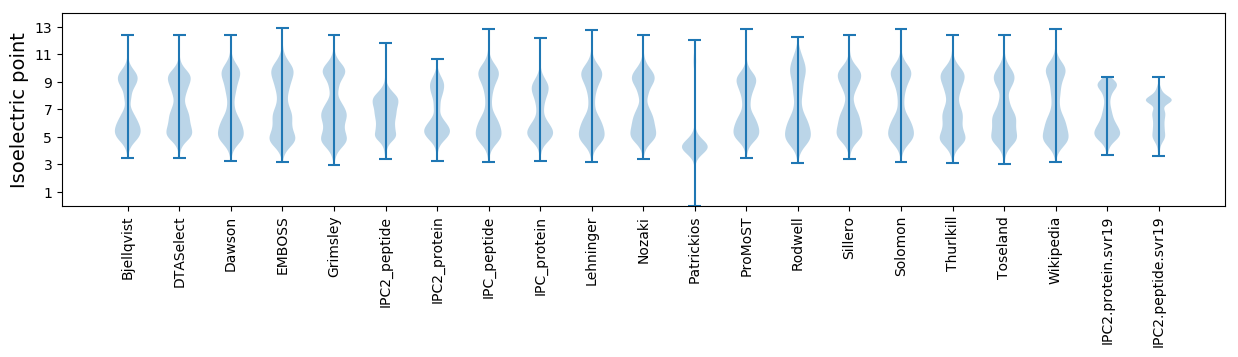
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M6A0A3|A0A1M6A0A3_9FLAO Glutamate--tRNA ligase OS=Arenibacter nanhaiticus OX=558155 GN=gltX PE=3 SV=1
MM1 pKa = 7.04TMYY4 pKa = 8.87RR5 pKa = 11.84TSYY8 pKa = 10.4EE9 pKa = 3.91VAQVLEE15 pKa = 4.38RR16 pKa = 11.84NRR18 pKa = 11.84EE19 pKa = 3.89QLAGLCANSWQSRR32 pKa = 11.84TLHH35 pKa = 6.62ALRR38 pKa = 11.84KK39 pKa = 9.5CRR41 pKa = 11.84TAALGGHH48 pKa = 7.01IDD50 pKa = 3.39RR51 pKa = 11.84CDD53 pKa = 3.33NPGCHH58 pKa = 6.95KK59 pKa = 10.79LHH61 pKa = 6.88LSYY64 pKa = 11.02NSCRR68 pKa = 11.84NRR70 pKa = 11.84HH71 pKa = 5.34CPKK74 pKa = 10.24CQGHH78 pKa = 6.77KK79 pKa = 9.45RR80 pKa = 11.84EE81 pKa = 3.69QWIRR85 pKa = 11.84ARR87 pKa = 11.84EE88 pKa = 4.01AEE90 pKa = 4.5LLNVTYY96 pKa = 10.04FHH98 pKa = 6.75MVFTLPSEE106 pKa = 4.28LNRR109 pKa = 11.84MCLYY113 pKa = 10.38DD114 pKa = 3.75PKK116 pKa = 10.77SIYY119 pKa = 10.89GLLFGTAWQVIKK131 pKa = 10.87GFSSNPKK138 pKa = 10.02FLGADD143 pKa = 3.32PGMVAILHH151 pKa = 6.06SWGQNLSLHH160 pKa = 6.08PHH162 pKa = 5.95LHH164 pKa = 6.6CIVPGGGTTRR174 pKa = 11.84SGKK177 pKa = 9.11WKK179 pKa = 9.77FARR182 pKa = 11.84NKK184 pKa = 9.96GKK186 pKa = 9.61PGCRR190 pKa = 11.84KK191 pKa = 9.43ARR193 pKa = 11.84YY194 pKa = 8.26LFPVKK199 pKa = 10.55AMSKK203 pKa = 8.25VFRR206 pKa = 11.84ARR208 pKa = 11.84FVASLRR214 pKa = 11.84KK215 pKa = 9.22EE216 pKa = 4.31FPPHH220 pKa = 5.26PTAFYY225 pKa = 11.01EE226 pKa = 4.95NLFKK230 pKa = 11.12HH231 pKa = 5.64NWVIYY236 pKa = 8.98CKK238 pKa = 10.32RR239 pKa = 11.84PFLGPEE245 pKa = 3.69QVVEE249 pKa = 3.68YY250 pKa = 10.34LGRR253 pKa = 11.84YY254 pKa = 3.88THH256 pKa = 7.36KK257 pKa = 10.47IAISNHH263 pKa = 5.96RR264 pKa = 11.84IKK266 pKa = 11.04SLDD269 pKa = 3.48NNNVTFTVKK278 pKa = 10.4DD279 pKa = 3.68YY280 pKa = 11.17RR281 pKa = 11.84HH282 pKa = 6.47GGRR285 pKa = 11.84KK286 pKa = 9.05SLLRR290 pKa = 11.84LTDD293 pKa = 3.21AEE295 pKa = 4.29FVRR298 pKa = 11.84RR299 pKa = 11.84FALHH303 pKa = 6.73ILPKK307 pKa = 8.86GFVRR311 pKa = 11.84IRR313 pKa = 11.84HH314 pKa = 5.2YY315 pKa = 11.2GILSSSRR322 pKa = 11.84KK323 pKa = 9.3KK324 pKa = 10.78ALLPLLQQVLGGPVPNGRR342 pKa = 11.84PPLKK346 pKa = 10.12HH347 pKa = 6.24RR348 pKa = 11.84RR349 pKa = 11.84CPYY352 pKa = 8.31CQIGIMVTVAVFDD365 pKa = 3.83SRR367 pKa = 11.84GPPKK371 pKa = 10.42AWSDD375 pKa = 3.52RR376 pKa = 11.84LKK378 pKa = 11.13NSAPRR383 pKa = 11.84RR384 pKa = 11.84TPQKK388 pKa = 10.74AA389 pKa = 2.8
MM1 pKa = 7.04TMYY4 pKa = 8.87RR5 pKa = 11.84TSYY8 pKa = 10.4EE9 pKa = 3.91VAQVLEE15 pKa = 4.38RR16 pKa = 11.84NRR18 pKa = 11.84EE19 pKa = 3.89QLAGLCANSWQSRR32 pKa = 11.84TLHH35 pKa = 6.62ALRR38 pKa = 11.84KK39 pKa = 9.5CRR41 pKa = 11.84TAALGGHH48 pKa = 7.01IDD50 pKa = 3.39RR51 pKa = 11.84CDD53 pKa = 3.33NPGCHH58 pKa = 6.95KK59 pKa = 10.79LHH61 pKa = 6.88LSYY64 pKa = 11.02NSCRR68 pKa = 11.84NRR70 pKa = 11.84HH71 pKa = 5.34CPKK74 pKa = 10.24CQGHH78 pKa = 6.77KK79 pKa = 9.45RR80 pKa = 11.84EE81 pKa = 3.69QWIRR85 pKa = 11.84ARR87 pKa = 11.84EE88 pKa = 4.01AEE90 pKa = 4.5LLNVTYY96 pKa = 10.04FHH98 pKa = 6.75MVFTLPSEE106 pKa = 4.28LNRR109 pKa = 11.84MCLYY113 pKa = 10.38DD114 pKa = 3.75PKK116 pKa = 10.77SIYY119 pKa = 10.89GLLFGTAWQVIKK131 pKa = 10.87GFSSNPKK138 pKa = 10.02FLGADD143 pKa = 3.32PGMVAILHH151 pKa = 6.06SWGQNLSLHH160 pKa = 6.08PHH162 pKa = 5.95LHH164 pKa = 6.6CIVPGGGTTRR174 pKa = 11.84SGKK177 pKa = 9.11WKK179 pKa = 9.77FARR182 pKa = 11.84NKK184 pKa = 9.96GKK186 pKa = 9.61PGCRR190 pKa = 11.84KK191 pKa = 9.43ARR193 pKa = 11.84YY194 pKa = 8.26LFPVKK199 pKa = 10.55AMSKK203 pKa = 8.25VFRR206 pKa = 11.84ARR208 pKa = 11.84FVASLRR214 pKa = 11.84KK215 pKa = 9.22EE216 pKa = 4.31FPPHH220 pKa = 5.26PTAFYY225 pKa = 11.01EE226 pKa = 4.95NLFKK230 pKa = 11.12HH231 pKa = 5.64NWVIYY236 pKa = 8.98CKK238 pKa = 10.32RR239 pKa = 11.84PFLGPEE245 pKa = 3.69QVVEE249 pKa = 3.68YY250 pKa = 10.34LGRR253 pKa = 11.84YY254 pKa = 3.88THH256 pKa = 7.36KK257 pKa = 10.47IAISNHH263 pKa = 5.96RR264 pKa = 11.84IKK266 pKa = 11.04SLDD269 pKa = 3.48NNNVTFTVKK278 pKa = 10.4DD279 pKa = 3.68YY280 pKa = 11.17RR281 pKa = 11.84HH282 pKa = 6.47GGRR285 pKa = 11.84KK286 pKa = 9.05SLLRR290 pKa = 11.84LTDD293 pKa = 3.21AEE295 pKa = 4.29FVRR298 pKa = 11.84RR299 pKa = 11.84FALHH303 pKa = 6.73ILPKK307 pKa = 8.86GFVRR311 pKa = 11.84IRR313 pKa = 11.84HH314 pKa = 5.2YY315 pKa = 11.2GILSSSRR322 pKa = 11.84KK323 pKa = 9.3KK324 pKa = 10.78ALLPLLQQVLGGPVPNGRR342 pKa = 11.84PPLKK346 pKa = 10.12HH347 pKa = 6.24RR348 pKa = 11.84RR349 pKa = 11.84CPYY352 pKa = 8.31CQIGIMVTVAVFDD365 pKa = 3.83SRR367 pKa = 11.84GPPKK371 pKa = 10.42AWSDD375 pKa = 3.52RR376 pKa = 11.84LKK378 pKa = 11.13NSAPRR383 pKa = 11.84RR384 pKa = 11.84TPQKK388 pKa = 10.74AA389 pKa = 2.8
Molecular weight: 44.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1230055 |
34 |
2809 |
346.2 |
38.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.643 ± 0.034 | 0.728 ± 0.012 |
5.5 ± 0.029 | 6.582 ± 0.037 |
5.077 ± 0.031 | 6.738 ± 0.043 |
1.905 ± 0.024 | 7.545 ± 0.035 |
7.606 ± 0.047 | 9.488 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.275 ± 0.02 | 5.786 ± 0.039 |
3.597 ± 0.024 | 3.434 ± 0.024 |
3.611 ± 0.024 | 6.437 ± 0.034 |
5.66 ± 0.044 | 6.195 ± 0.034 |
1.141 ± 0.018 | 4.055 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
