
Streptomyces cyaneogriseus subsp. noncyanogenus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; Streptomyces cyaneogriseus
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
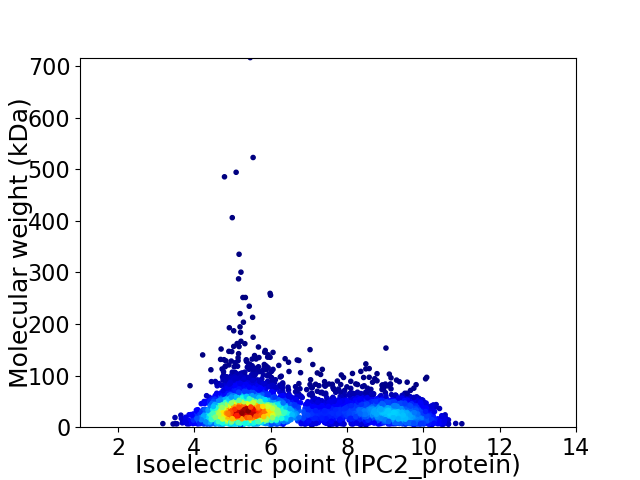
Virtual 2D-PAGE plot for 5829 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5FX28|A0A0C5FX28_9ACTN Uncharacterized protein OS=Streptomyces cyaneogriseus subsp. noncyanogenus OX=477245 GN=TU94_12950 PE=4 SV=1
MM1 pKa = 7.83TDD3 pKa = 3.35DD4 pKa = 3.32QHH6 pKa = 9.03AEE8 pKa = 3.86GMGQGGGRR16 pKa = 11.84WDD18 pKa = 4.35PLPQGDD24 pKa = 4.33YY25 pKa = 11.28DD26 pKa = 4.37DD27 pKa = 4.93GATAFVKK34 pKa = 10.6LPEE37 pKa = 4.6GGIDD41 pKa = 3.79ALLASADD48 pKa = 3.98SPLAAPGHH56 pKa = 6.36GYY58 pKa = 9.65VPPRR62 pKa = 11.84ITVAPTTPAATDD74 pKa = 3.65PAAPGPWGAPDD85 pKa = 5.81GGAQWHH91 pKa = 6.68APDD94 pKa = 4.6APEE97 pKa = 4.95GGYY100 pKa = 8.76PHH102 pKa = 7.14QDD104 pKa = 2.45TGYY107 pKa = 9.15GQHH110 pKa = 6.25GQDD113 pKa = 3.73AGYY116 pKa = 8.59PQDD119 pKa = 3.38GGYY122 pKa = 10.43AQGGDD127 pKa = 3.27GGYY130 pKa = 9.04PHH132 pKa = 6.99QGQDD136 pKa = 2.85AGYY139 pKa = 8.33GQHH142 pKa = 6.55GRR144 pKa = 11.84DD145 pKa = 2.92AGYY148 pKa = 8.94PQGFDD153 pKa = 2.87GGYY156 pKa = 9.52EE157 pKa = 3.83QDD159 pKa = 3.61PEE161 pKa = 4.31AGYY164 pKa = 10.19RR165 pKa = 11.84QGEE168 pKa = 4.35DD169 pKa = 2.29AGYY172 pKa = 9.27PYY174 pKa = 10.46GAPAGYY180 pKa = 9.14RR181 pKa = 11.84QHH183 pKa = 7.23DD184 pKa = 4.2PYY186 pKa = 11.43GVAGRR191 pKa = 11.84QDD193 pKa = 3.22GAQYY197 pKa = 10.63DD198 pKa = 4.02PHH200 pKa = 8.07AGPQYY205 pKa = 10.25VADD208 pKa = 4.72DD209 pKa = 3.93RR210 pKa = 11.84FTYY213 pKa = 10.45HH214 pKa = 7.33PGATGQWDD222 pKa = 3.84YY223 pKa = 11.81TEE225 pKa = 5.31AAAQEE230 pKa = 4.25DD231 pKa = 4.38TAAAAPAPAPGHH243 pKa = 6.77DD244 pKa = 3.67VTGQWSIPVAGGDD257 pKa = 3.84LPDD260 pKa = 3.81EE261 pKa = 4.7SGEE264 pKa = 4.14FTTSSLVEE272 pKa = 4.05QWGGTPPATLPGGASAPWAPPAAGPQAHH300 pKa = 6.89GDD302 pKa = 3.59VDD304 pKa = 3.89PAADD308 pKa = 3.76PFAGHH313 pKa = 7.39GGRR316 pKa = 11.84QDD318 pKa = 3.86DD319 pKa = 4.1APVAPEE325 pKa = 4.55HH326 pKa = 6.61GFAPYY331 pKa = 10.84AEE333 pKa = 4.74DD334 pKa = 3.17AGEE337 pKa = 4.09AVEE340 pKa = 4.33QPEE343 pKa = 4.77PEE345 pKa = 4.21PEE347 pKa = 4.49GPGVPQEE354 pKa = 4.37PGQEE358 pKa = 4.14SEE360 pKa = 5.05APGTPEE366 pKa = 3.94GQLPAQDD373 pKa = 3.53SVEE376 pKa = 4.16PAEE379 pKa = 4.43EE380 pKa = 4.09TSPPAAAPEE389 pKa = 4.3APEE392 pKa = 4.39GWQGAEE398 pKa = 4.13QPDD401 pKa = 4.46GSPEE405 pKa = 4.22TPSEE409 pKa = 4.1AQDD412 pKa = 3.8GPEE415 pKa = 4.39APSPAQDD422 pKa = 3.34APEE425 pKa = 3.98AAEE428 pKa = 4.18AQDD431 pKa = 3.58AVEE434 pKa = 5.05DD435 pKa = 4.07VPEE438 pKa = 4.33ATDD441 pKa = 3.85APDD444 pKa = 3.25MPEE447 pKa = 4.17DD448 pKa = 3.75SGAGEE453 pKa = 4.34PLDD456 pKa = 4.97ARR458 pKa = 11.84DD459 pKa = 3.75GQEE462 pKa = 3.8AQHH465 pKa = 6.27AHH467 pKa = 7.19DD468 pKa = 4.75GQHH471 pKa = 7.03AYY473 pKa = 10.13DD474 pKa = 4.06AQHH477 pKa = 6.95APGEE481 pKa = 4.36HH482 pKa = 6.89DD483 pKa = 4.39AVPAPEE489 pKa = 5.03GPAEE493 pKa = 4.3PFAAPAAAHH502 pKa = 6.83DD503 pKa = 4.1AQAAPEE509 pKa = 4.2AEE511 pKa = 4.43EE512 pKa = 4.91APEE515 pKa = 4.06APEE518 pKa = 3.83TSEE521 pKa = 3.96TSGTSEE527 pKa = 4.03TPEE530 pKa = 4.02AATGPGAPADD540 pKa = 4.11PADD543 pKa = 4.63GPDD546 pKa = 3.64PAAGTDD552 pKa = 3.63PADD555 pKa = 4.25PSAGTAPAPGDD566 pKa = 4.09EE567 pKa = 4.72PPAGAPAEE575 pKa = 4.26PEE577 pKa = 4.24EE578 pKa = 4.52EE579 pKa = 4.09PAAAPAAHH587 pKa = 7.33DD588 pKa = 3.78EE589 pKa = 4.64HH590 pKa = 7.23PLASYY595 pKa = 9.26VLRR598 pKa = 11.84VNGTDD603 pKa = 4.45RR604 pKa = 11.84PVTDD608 pKa = 2.98AWIGEE613 pKa = 4.15SLLYY617 pKa = 10.18VLRR620 pKa = 11.84EE621 pKa = 3.94RR622 pKa = 11.84LGLAGAKK629 pKa = 9.72DD630 pKa = 3.75GCSQGEE636 pKa = 4.12CGACNVQVDD645 pKa = 3.94GRR647 pKa = 11.84LVASCLVPAATAAGSEE663 pKa = 4.08VRR665 pKa = 11.84TVEE668 pKa = 4.06GLAADD673 pKa = 4.65GQPSDD678 pKa = 3.75VQRR681 pKa = 11.84ALARR685 pKa = 11.84CGAVQCGFCVPGMAMTVHH703 pKa = 7.27DD704 pKa = 5.16LLEE707 pKa = 4.78GNPAPTEE714 pKa = 4.03LEE716 pKa = 3.87ARR718 pKa = 11.84QALCGNLCRR727 pKa = 11.84CSGYY731 pKa = 10.32RR732 pKa = 11.84GVLRR736 pKa = 11.84AVEE739 pKa = 4.13EE740 pKa = 4.62VVAEE744 pKa = 4.72RR745 pKa = 11.84GAHH748 pKa = 5.08SAADD752 pKa = 3.47ASEE755 pKa = 4.32ADD757 pKa = 3.68GGRR760 pKa = 11.84TRR762 pKa = 11.84VPHH765 pKa = 5.19QAGPEE770 pKa = 3.86AGGAEE775 pKa = 4.57PSPFDD780 pKa = 3.93PPEE783 pKa = 4.81APGPHH788 pKa = 6.54DD789 pKa = 3.5QPYY792 pKa = 9.54GQDD795 pKa = 3.28GGHH798 pKa = 6.65AA799 pKa = 3.82
MM1 pKa = 7.83TDD3 pKa = 3.35DD4 pKa = 3.32QHH6 pKa = 9.03AEE8 pKa = 3.86GMGQGGGRR16 pKa = 11.84WDD18 pKa = 4.35PLPQGDD24 pKa = 4.33YY25 pKa = 11.28DD26 pKa = 4.37DD27 pKa = 4.93GATAFVKK34 pKa = 10.6LPEE37 pKa = 4.6GGIDD41 pKa = 3.79ALLASADD48 pKa = 3.98SPLAAPGHH56 pKa = 6.36GYY58 pKa = 9.65VPPRR62 pKa = 11.84ITVAPTTPAATDD74 pKa = 3.65PAAPGPWGAPDD85 pKa = 5.81GGAQWHH91 pKa = 6.68APDD94 pKa = 4.6APEE97 pKa = 4.95GGYY100 pKa = 8.76PHH102 pKa = 7.14QDD104 pKa = 2.45TGYY107 pKa = 9.15GQHH110 pKa = 6.25GQDD113 pKa = 3.73AGYY116 pKa = 8.59PQDD119 pKa = 3.38GGYY122 pKa = 10.43AQGGDD127 pKa = 3.27GGYY130 pKa = 9.04PHH132 pKa = 6.99QGQDD136 pKa = 2.85AGYY139 pKa = 8.33GQHH142 pKa = 6.55GRR144 pKa = 11.84DD145 pKa = 2.92AGYY148 pKa = 8.94PQGFDD153 pKa = 2.87GGYY156 pKa = 9.52EE157 pKa = 3.83QDD159 pKa = 3.61PEE161 pKa = 4.31AGYY164 pKa = 10.19RR165 pKa = 11.84QGEE168 pKa = 4.35DD169 pKa = 2.29AGYY172 pKa = 9.27PYY174 pKa = 10.46GAPAGYY180 pKa = 9.14RR181 pKa = 11.84QHH183 pKa = 7.23DD184 pKa = 4.2PYY186 pKa = 11.43GVAGRR191 pKa = 11.84QDD193 pKa = 3.22GAQYY197 pKa = 10.63DD198 pKa = 4.02PHH200 pKa = 8.07AGPQYY205 pKa = 10.25VADD208 pKa = 4.72DD209 pKa = 3.93RR210 pKa = 11.84FTYY213 pKa = 10.45HH214 pKa = 7.33PGATGQWDD222 pKa = 3.84YY223 pKa = 11.81TEE225 pKa = 5.31AAAQEE230 pKa = 4.25DD231 pKa = 4.38TAAAAPAPAPGHH243 pKa = 6.77DD244 pKa = 3.67VTGQWSIPVAGGDD257 pKa = 3.84LPDD260 pKa = 3.81EE261 pKa = 4.7SGEE264 pKa = 4.14FTTSSLVEE272 pKa = 4.05QWGGTPPATLPGGASAPWAPPAAGPQAHH300 pKa = 6.89GDD302 pKa = 3.59VDD304 pKa = 3.89PAADD308 pKa = 3.76PFAGHH313 pKa = 7.39GGRR316 pKa = 11.84QDD318 pKa = 3.86DD319 pKa = 4.1APVAPEE325 pKa = 4.55HH326 pKa = 6.61GFAPYY331 pKa = 10.84AEE333 pKa = 4.74DD334 pKa = 3.17AGEE337 pKa = 4.09AVEE340 pKa = 4.33QPEE343 pKa = 4.77PEE345 pKa = 4.21PEE347 pKa = 4.49GPGVPQEE354 pKa = 4.37PGQEE358 pKa = 4.14SEE360 pKa = 5.05APGTPEE366 pKa = 3.94GQLPAQDD373 pKa = 3.53SVEE376 pKa = 4.16PAEE379 pKa = 4.43EE380 pKa = 4.09TSPPAAAPEE389 pKa = 4.3APEE392 pKa = 4.39GWQGAEE398 pKa = 4.13QPDD401 pKa = 4.46GSPEE405 pKa = 4.22TPSEE409 pKa = 4.1AQDD412 pKa = 3.8GPEE415 pKa = 4.39APSPAQDD422 pKa = 3.34APEE425 pKa = 3.98AAEE428 pKa = 4.18AQDD431 pKa = 3.58AVEE434 pKa = 5.05DD435 pKa = 4.07VPEE438 pKa = 4.33ATDD441 pKa = 3.85APDD444 pKa = 3.25MPEE447 pKa = 4.17DD448 pKa = 3.75SGAGEE453 pKa = 4.34PLDD456 pKa = 4.97ARR458 pKa = 11.84DD459 pKa = 3.75GQEE462 pKa = 3.8AQHH465 pKa = 6.27AHH467 pKa = 7.19DD468 pKa = 4.75GQHH471 pKa = 7.03AYY473 pKa = 10.13DD474 pKa = 4.06AQHH477 pKa = 6.95APGEE481 pKa = 4.36HH482 pKa = 6.89DD483 pKa = 4.39AVPAPEE489 pKa = 5.03GPAEE493 pKa = 4.3PFAAPAAAHH502 pKa = 6.83DD503 pKa = 4.1AQAAPEE509 pKa = 4.2AEE511 pKa = 4.43EE512 pKa = 4.91APEE515 pKa = 4.06APEE518 pKa = 3.83TSEE521 pKa = 3.96TSGTSEE527 pKa = 4.03TPEE530 pKa = 4.02AATGPGAPADD540 pKa = 4.11PADD543 pKa = 4.63GPDD546 pKa = 3.64PAAGTDD552 pKa = 3.63PADD555 pKa = 4.25PSAGTAPAPGDD566 pKa = 4.09EE567 pKa = 4.72PPAGAPAEE575 pKa = 4.26PEE577 pKa = 4.24EE578 pKa = 4.52EE579 pKa = 4.09PAAAPAAHH587 pKa = 7.33DD588 pKa = 3.78EE589 pKa = 4.64HH590 pKa = 7.23PLASYY595 pKa = 9.26VLRR598 pKa = 11.84VNGTDD603 pKa = 4.45RR604 pKa = 11.84PVTDD608 pKa = 2.98AWIGEE613 pKa = 4.15SLLYY617 pKa = 10.18VLRR620 pKa = 11.84EE621 pKa = 3.94RR622 pKa = 11.84LGLAGAKK629 pKa = 9.72DD630 pKa = 3.75GCSQGEE636 pKa = 4.12CGACNVQVDD645 pKa = 3.94GRR647 pKa = 11.84LVASCLVPAATAAGSEE663 pKa = 4.08VRR665 pKa = 11.84TVEE668 pKa = 4.06GLAADD673 pKa = 4.65GQPSDD678 pKa = 3.75VQRR681 pKa = 11.84ALARR685 pKa = 11.84CGAVQCGFCVPGMAMTVHH703 pKa = 7.27DD704 pKa = 5.16LLEE707 pKa = 4.78GNPAPTEE714 pKa = 4.03LEE716 pKa = 3.87ARR718 pKa = 11.84QALCGNLCRR727 pKa = 11.84CSGYY731 pKa = 10.32RR732 pKa = 11.84GVLRR736 pKa = 11.84AVEE739 pKa = 4.13EE740 pKa = 4.62VVAEE744 pKa = 4.72RR745 pKa = 11.84GAHH748 pKa = 5.08SAADD752 pKa = 3.47ASEE755 pKa = 4.32ADD757 pKa = 3.68GGRR760 pKa = 11.84TRR762 pKa = 11.84VPHH765 pKa = 5.19QAGPEE770 pKa = 3.86AGGAEE775 pKa = 4.57PSPFDD780 pKa = 3.93PPEE783 pKa = 4.81APGPHH788 pKa = 6.54DD789 pKa = 3.5QPYY792 pKa = 9.54GQDD795 pKa = 3.28GGHH798 pKa = 6.65AA799 pKa = 3.82
Molecular weight: 80.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5FVJ8|A0A0C5FVJ8_9ACTN Regulatory protein OS=Streptomyces cyaneogriseus subsp. noncyanogenus OX=477245 GN=TU94_09590 PE=4 SV=1
MM1 pKa = 7.47ARR3 pKa = 11.84TAVRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84PPAPYY14 pKa = 9.67RR15 pKa = 11.84RR16 pKa = 11.84PAPALRR22 pKa = 11.84PSPPRR27 pKa = 11.84PARR30 pKa = 11.84ARR32 pKa = 11.84PGRR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84ARR40 pKa = 11.84GQPSPACRR48 pKa = 11.84PGVRR52 pKa = 11.84TSSSTLSTVSS62 pKa = 3.01
MM1 pKa = 7.47ARR3 pKa = 11.84TAVRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84PPAPYY14 pKa = 9.67RR15 pKa = 11.84RR16 pKa = 11.84PAPALRR22 pKa = 11.84PSPPRR27 pKa = 11.84PARR30 pKa = 11.84ARR32 pKa = 11.84PGRR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84ARR40 pKa = 11.84GQPSPACRR48 pKa = 11.84PGVRR52 pKa = 11.84TSSSTLSTVSS62 pKa = 3.01
Molecular weight: 6.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1944252 |
29 |
6673 |
333.5 |
35.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.983 ± 0.053 | 0.794 ± 0.008 |
5.944 ± 0.026 | 5.881 ± 0.035 |
2.683 ± 0.019 | 9.652 ± 0.038 |
2.305 ± 0.015 | 2.999 ± 0.023 |
2.053 ± 0.029 | 10.278 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.685 ± 0.014 | 1.639 ± 0.016 |
6.228 ± 0.033 | 2.621 ± 0.018 |
8.452 ± 0.041 | 4.673 ± 0.025 |
6.015 ± 0.026 | 8.492 ± 0.031 |
1.509 ± 0.016 | 2.112 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
