
Changjiang tombus-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.52
Get precalculated fractions of proteins
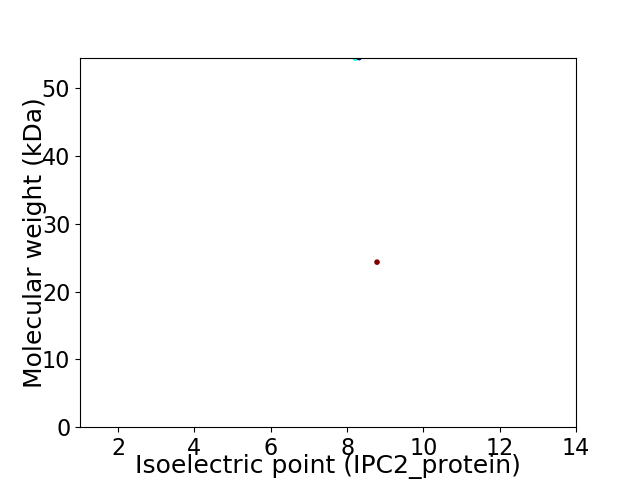
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KG78|A0A1L3KG78_9VIRU RNA-directed RNA polymerase OS=Changjiang tombus-like virus 2 OX=1922813 PE=4 SV=1
MM1 pKa = 7.06GQHH4 pKa = 6.54IVDD7 pKa = 4.95DD8 pKa = 4.15NLNTHH13 pKa = 6.12NNSVANLLRR22 pKa = 11.84GVGEE26 pKa = 3.95RR27 pKa = 11.84VLYY30 pKa = 9.82TDD32 pKa = 5.22RR33 pKa = 11.84NLTKK37 pKa = 10.4CAQPLSGVFEE47 pKa = 4.47GRR49 pKa = 11.84LASYY53 pKa = 9.86KK54 pKa = 10.07RR55 pKa = 11.84RR56 pKa = 11.84IVRR59 pKa = 11.84SVGRR63 pKa = 11.84QSPVTRR69 pKa = 11.84DD70 pKa = 3.31QFVEE74 pKa = 5.2FYY76 pKa = 10.17KK77 pKa = 10.69GPRR80 pKa = 11.84RR81 pKa = 11.84ATYY84 pKa = 9.07EE85 pKa = 3.78RR86 pKa = 11.84AVASLVLKK94 pKa = 7.98PICSRR99 pKa = 11.84DD100 pKa = 3.29ARR102 pKa = 11.84LKK104 pKa = 9.98TFVKK108 pKa = 10.45AEE110 pKa = 4.41KK111 pKa = 10.46INFTLKK117 pKa = 10.21EE118 pKa = 4.11DD119 pKa = 3.76PAPRR123 pKa = 11.84VIQPRR128 pKa = 11.84EE129 pKa = 3.41PRR131 pKa = 11.84FNVEE135 pKa = 3.33VGKK138 pKa = 9.05YY139 pKa = 9.43LRR141 pKa = 11.84PIEE144 pKa = 4.45HH145 pKa = 6.8KK146 pKa = 10.92VYY148 pKa = 10.89DD149 pKa = 5.2AIDD152 pKa = 4.07DD153 pKa = 4.32LFGSPTVMSKK163 pKa = 10.99YY164 pKa = 10.66NSVQTANIIHH174 pKa = 7.2DD175 pKa = 4.04KK176 pKa = 10.14FSSIFGCAVVGLDD189 pKa = 4.03ASRR192 pKa = 11.84FDD194 pKa = 3.47QHH196 pKa = 8.35VSEE199 pKa = 4.21QALKK203 pKa = 10.24FEE205 pKa = 4.33HH206 pKa = 6.77SIYY209 pKa = 10.91DD210 pKa = 3.85GIFNSGEE217 pKa = 3.9LRR219 pKa = 11.84WLLKK223 pKa = 10.46HH224 pKa = 5.83QLHH227 pKa = 5.88NHH229 pKa = 5.8GFAKK233 pKa = 10.86GNDD236 pKa = 2.99GWFKK240 pKa = 9.53YY241 pKa = 8.82QKK243 pKa = 9.88KK244 pKa = 8.61GSRR247 pKa = 11.84MSGDD251 pKa = 3.39MNTSLGNKK259 pKa = 9.73LLMCMMCKK267 pKa = 10.2SYY269 pKa = 11.11LDD271 pKa = 3.72SLKK274 pKa = 10.4IPYY277 pKa = 10.02EE278 pKa = 4.01FVNNGDD284 pKa = 3.6DD285 pKa = 3.64CLVFISRR292 pKa = 11.84KK293 pKa = 9.32HH294 pKa = 5.18LPKK297 pKa = 10.71LEE299 pKa = 4.15TLDD302 pKa = 4.61QYY304 pKa = 11.7FKK306 pKa = 11.52DD307 pKa = 4.17FGFKK311 pKa = 9.22MKK313 pKa = 10.59CEE315 pKa = 3.95PPVFEE320 pKa = 4.24VEE322 pKa = 4.18QIEE325 pKa = 4.59FCQCKK330 pKa = 9.23PVLCNGIWRR339 pKa = 11.84MTRR342 pKa = 11.84NIRR345 pKa = 11.84TALAKK350 pKa = 10.61DD351 pKa = 3.74CTSVNLGHH359 pKa = 7.46DD360 pKa = 3.27IEE362 pKa = 4.93LFRR365 pKa = 11.84RR366 pKa = 11.84WLHH369 pKa = 6.66DD370 pKa = 3.37VSACGAAFSADD381 pKa = 3.75LPVLGSFYY389 pKa = 11.18RR390 pKa = 11.84MLGRR394 pKa = 11.84FGIAGEE400 pKa = 4.32YY401 pKa = 9.6EE402 pKa = 4.11GHH404 pKa = 6.35KK405 pKa = 10.63SEE407 pKa = 4.28FAAYY411 pKa = 9.59RR412 pKa = 11.84SMSRR416 pKa = 11.84GVHH419 pKa = 5.33IPYY422 pKa = 9.69NAPNAQGRR430 pKa = 11.84YY431 pKa = 9.19SFWLSTGINPEE442 pKa = 3.68QQEE445 pKa = 4.07IIEE448 pKa = 4.7NYY450 pKa = 9.79FDD452 pKa = 3.5TAVWGGDD459 pKa = 2.64KK460 pKa = 10.59RR461 pKa = 11.84QIINSIDD468 pKa = 3.45YY469 pKa = 10.57IITHH473 pKa = 6.01GRR475 pKa = 11.84NN476 pKa = 3.04
MM1 pKa = 7.06GQHH4 pKa = 6.54IVDD7 pKa = 4.95DD8 pKa = 4.15NLNTHH13 pKa = 6.12NNSVANLLRR22 pKa = 11.84GVGEE26 pKa = 3.95RR27 pKa = 11.84VLYY30 pKa = 9.82TDD32 pKa = 5.22RR33 pKa = 11.84NLTKK37 pKa = 10.4CAQPLSGVFEE47 pKa = 4.47GRR49 pKa = 11.84LASYY53 pKa = 9.86KK54 pKa = 10.07RR55 pKa = 11.84RR56 pKa = 11.84IVRR59 pKa = 11.84SVGRR63 pKa = 11.84QSPVTRR69 pKa = 11.84DD70 pKa = 3.31QFVEE74 pKa = 5.2FYY76 pKa = 10.17KK77 pKa = 10.69GPRR80 pKa = 11.84RR81 pKa = 11.84ATYY84 pKa = 9.07EE85 pKa = 3.78RR86 pKa = 11.84AVASLVLKK94 pKa = 7.98PICSRR99 pKa = 11.84DD100 pKa = 3.29ARR102 pKa = 11.84LKK104 pKa = 9.98TFVKK108 pKa = 10.45AEE110 pKa = 4.41KK111 pKa = 10.46INFTLKK117 pKa = 10.21EE118 pKa = 4.11DD119 pKa = 3.76PAPRR123 pKa = 11.84VIQPRR128 pKa = 11.84EE129 pKa = 3.41PRR131 pKa = 11.84FNVEE135 pKa = 3.33VGKK138 pKa = 9.05YY139 pKa = 9.43LRR141 pKa = 11.84PIEE144 pKa = 4.45HH145 pKa = 6.8KK146 pKa = 10.92VYY148 pKa = 10.89DD149 pKa = 5.2AIDD152 pKa = 4.07DD153 pKa = 4.32LFGSPTVMSKK163 pKa = 10.99YY164 pKa = 10.66NSVQTANIIHH174 pKa = 7.2DD175 pKa = 4.04KK176 pKa = 10.14FSSIFGCAVVGLDD189 pKa = 4.03ASRR192 pKa = 11.84FDD194 pKa = 3.47QHH196 pKa = 8.35VSEE199 pKa = 4.21QALKK203 pKa = 10.24FEE205 pKa = 4.33HH206 pKa = 6.77SIYY209 pKa = 10.91DD210 pKa = 3.85GIFNSGEE217 pKa = 3.9LRR219 pKa = 11.84WLLKK223 pKa = 10.46HH224 pKa = 5.83QLHH227 pKa = 5.88NHH229 pKa = 5.8GFAKK233 pKa = 10.86GNDD236 pKa = 2.99GWFKK240 pKa = 9.53YY241 pKa = 8.82QKK243 pKa = 9.88KK244 pKa = 8.61GSRR247 pKa = 11.84MSGDD251 pKa = 3.39MNTSLGNKK259 pKa = 9.73LLMCMMCKK267 pKa = 10.2SYY269 pKa = 11.11LDD271 pKa = 3.72SLKK274 pKa = 10.4IPYY277 pKa = 10.02EE278 pKa = 4.01FVNNGDD284 pKa = 3.6DD285 pKa = 3.64CLVFISRR292 pKa = 11.84KK293 pKa = 9.32HH294 pKa = 5.18LPKK297 pKa = 10.71LEE299 pKa = 4.15TLDD302 pKa = 4.61QYY304 pKa = 11.7FKK306 pKa = 11.52DD307 pKa = 4.17FGFKK311 pKa = 9.22MKK313 pKa = 10.59CEE315 pKa = 3.95PPVFEE320 pKa = 4.24VEE322 pKa = 4.18QIEE325 pKa = 4.59FCQCKK330 pKa = 9.23PVLCNGIWRR339 pKa = 11.84MTRR342 pKa = 11.84NIRR345 pKa = 11.84TALAKK350 pKa = 10.61DD351 pKa = 3.74CTSVNLGHH359 pKa = 7.46DD360 pKa = 3.27IEE362 pKa = 4.93LFRR365 pKa = 11.84RR366 pKa = 11.84WLHH369 pKa = 6.66DD370 pKa = 3.37VSACGAAFSADD381 pKa = 3.75LPVLGSFYY389 pKa = 11.18RR390 pKa = 11.84MLGRR394 pKa = 11.84FGIAGEE400 pKa = 4.32YY401 pKa = 9.6EE402 pKa = 4.11GHH404 pKa = 6.35KK405 pKa = 10.63SEE407 pKa = 4.28FAAYY411 pKa = 9.59RR412 pKa = 11.84SMSRR416 pKa = 11.84GVHH419 pKa = 5.33IPYY422 pKa = 9.69NAPNAQGRR430 pKa = 11.84YY431 pKa = 9.19SFWLSTGINPEE442 pKa = 3.68QQEE445 pKa = 4.07IIEE448 pKa = 4.7NYY450 pKa = 9.79FDD452 pKa = 3.5TAVWGGDD459 pKa = 2.64KK460 pKa = 10.59RR461 pKa = 11.84QIINSIDD468 pKa = 3.45YY469 pKa = 10.57IITHH473 pKa = 6.01GRR475 pKa = 11.84NN476 pKa = 3.04
Molecular weight: 54.43 kDa
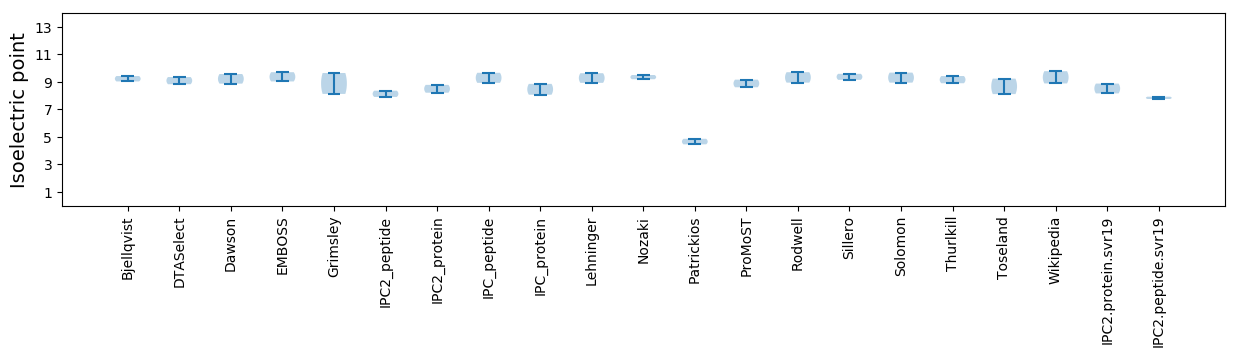
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KG78|A0A1L3KG78_9VIRU RNA-directed RNA polymerase OS=Changjiang tombus-like virus 2 OX=1922813 PE=4 SV=1
MM1 pKa = 7.79AGTKK5 pKa = 8.78RR6 pKa = 11.84TQRR9 pKa = 11.84TARR12 pKa = 11.84EE13 pKa = 4.02TVDD16 pKa = 3.09QHH18 pKa = 5.51SQRR21 pKa = 11.84SPRR24 pKa = 11.84VNLRR28 pKa = 11.84ARR30 pKa = 11.84LNEE33 pKa = 3.79NVSYY37 pKa = 10.97ISGNEE42 pKa = 3.9YY43 pKa = 8.79NTGAIVPSGSSLGYY57 pKa = 10.39GSVSLAPGNLAGRR70 pKa = 11.84ANAAINGVGRR80 pKa = 11.84FFQKK84 pKa = 10.72GLYY87 pKa = 10.32LPGTFVRR94 pKa = 11.84YY95 pKa = 9.35IPSVGLNTPGNIIIAWLDD113 pKa = 3.49NPDD116 pKa = 5.03MIRR119 pKa = 11.84AWNLLSAGAHH129 pKa = 5.98LNFIRR134 pKa = 11.84DD135 pKa = 3.37VSNAKK140 pKa = 9.06TGPVWQEE147 pKa = 3.3LTVPLTQPPRR157 pKa = 11.84RR158 pKa = 11.84KK159 pKa = 8.3TFMVDD164 pKa = 3.01PQLNFQSNTEE174 pKa = 3.87VDD176 pKa = 4.62LSCQGLFIFCVFGTDD191 pKa = 4.53ISLADD196 pKa = 4.12DD197 pKa = 3.44KK198 pKa = 11.45TYY200 pKa = 11.01GQLLVHH206 pKa = 6.51CKK208 pKa = 8.87MRR210 pKa = 11.84FEE212 pKa = 4.24EE213 pKa = 4.43VKK215 pKa = 11.01SFVTPQQ221 pKa = 2.73
MM1 pKa = 7.79AGTKK5 pKa = 8.78RR6 pKa = 11.84TQRR9 pKa = 11.84TARR12 pKa = 11.84EE13 pKa = 4.02TVDD16 pKa = 3.09QHH18 pKa = 5.51SQRR21 pKa = 11.84SPRR24 pKa = 11.84VNLRR28 pKa = 11.84ARR30 pKa = 11.84LNEE33 pKa = 3.79NVSYY37 pKa = 10.97ISGNEE42 pKa = 3.9YY43 pKa = 8.79NTGAIVPSGSSLGYY57 pKa = 10.39GSVSLAPGNLAGRR70 pKa = 11.84ANAAINGVGRR80 pKa = 11.84FFQKK84 pKa = 10.72GLYY87 pKa = 10.32LPGTFVRR94 pKa = 11.84YY95 pKa = 9.35IPSVGLNTPGNIIIAWLDD113 pKa = 3.49NPDD116 pKa = 5.03MIRR119 pKa = 11.84AWNLLSAGAHH129 pKa = 5.98LNFIRR134 pKa = 11.84DD135 pKa = 3.37VSNAKK140 pKa = 9.06TGPVWQEE147 pKa = 3.3LTVPLTQPPRR157 pKa = 11.84RR158 pKa = 11.84KK159 pKa = 8.3TFMVDD164 pKa = 3.01PQLNFQSNTEE174 pKa = 3.87VDD176 pKa = 4.62LSCQGLFIFCVFGTDD191 pKa = 4.53ISLADD196 pKa = 4.12DD197 pKa = 3.44KK198 pKa = 11.45TYY200 pKa = 11.01GQLLVHH206 pKa = 6.51CKK208 pKa = 8.87MRR210 pKa = 11.84FEE212 pKa = 4.24EE213 pKa = 4.43VKK215 pKa = 11.01SFVTPQQ221 pKa = 2.73
Molecular weight: 24.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
697 |
221 |
476 |
348.5 |
39.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.026 ± 0.364 | 2.152 ± 0.38 |
5.165 ± 0.522 | 4.591 ± 0.68 |
5.595 ± 0.295 | 7.747 ± 0.406 |
2.582 ± 0.586 | 5.595 ± 0.295 |
5.308 ± 1.023 | 8.178 ± 0.417 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.152 ± 0.164 | 6.026 ± 0.58 |
4.591 ± 0.617 | 4.017 ± 0.459 |
7.03 ± 0.116 | 6.887 ± 0.169 |
4.591 ± 1.05 | 6.887 ± 0.385 |
1.291 ± 0.032 | 3.587 ± 0.417 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
