
Gallionella capsiferriformans (strain ES-2) (Gallionella ferruginea capsiferriformans (strain ES-2))
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Nitrosomonadales; Gallionellaceae; Gallionella; Gallionella capsiferriformans
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
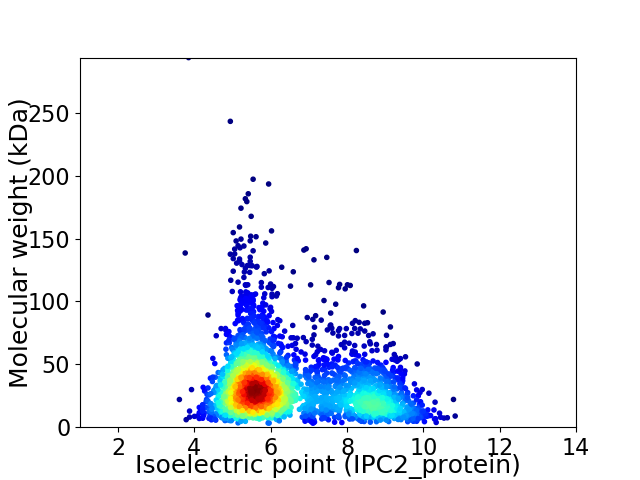
Virtual 2D-PAGE plot for 2853 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
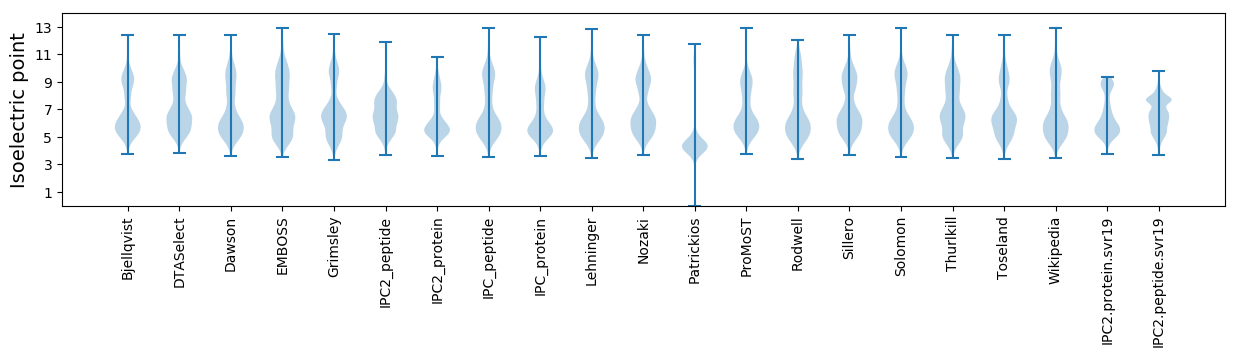
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D9SJS4|D9SJS4_GALCS Site-specific DNA-methyltransferase (adenine-specific) OS=Gallionella capsiferriformans (strain ES-2) OX=395494 GN=Galf_0379 PE=3 SV=1
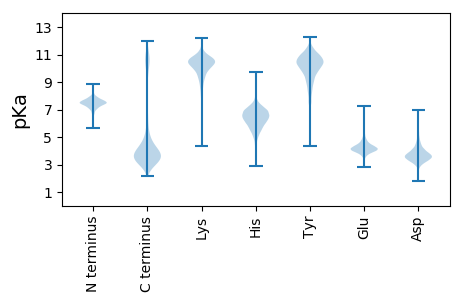
MM1 pKa = 8.1DD2 pKa = 3.91ATTAVTEE9 pKa = 4.31SQDD12 pKa = 3.88LLVFTDD18 pKa = 3.4NAAAKK23 pKa = 9.25VKK25 pKa = 10.37QLIEE29 pKa = 4.06EE30 pKa = 4.36EE31 pKa = 4.67GNSEE35 pKa = 3.75LKK37 pKa = 10.89LRR39 pKa = 11.84VFVSGGGCSGFQYY52 pKa = 10.94GFTFDD57 pKa = 4.81EE58 pKa = 4.64ITNEE62 pKa = 4.19DD63 pKa = 3.91DD64 pKa = 3.99TVLDD68 pKa = 3.86KK69 pKa = 11.81NGVQLLIDD77 pKa = 3.92PMSYY81 pKa = 10.31QYY83 pKa = 11.58LVGAEE88 pKa = 3.64IDD90 pKa = 3.75YY91 pKa = 10.77TEE93 pKa = 4.43GLEE96 pKa = 4.39GSQFVIKK103 pKa = 10.58NPNATTTCGCGSSFSVV119 pKa = 3.54
MM1 pKa = 8.1DD2 pKa = 3.91ATTAVTEE9 pKa = 4.31SQDD12 pKa = 3.88LLVFTDD18 pKa = 3.4NAAAKK23 pKa = 9.25VKK25 pKa = 10.37QLIEE29 pKa = 4.06EE30 pKa = 4.36EE31 pKa = 4.67GNSEE35 pKa = 3.75LKK37 pKa = 10.89LRR39 pKa = 11.84VFVSGGGCSGFQYY52 pKa = 10.94GFTFDD57 pKa = 4.81EE58 pKa = 4.64ITNEE62 pKa = 4.19DD63 pKa = 3.91DD64 pKa = 3.99TVLDD68 pKa = 3.86KK69 pKa = 11.81NGVQLLIDD77 pKa = 3.92PMSYY81 pKa = 10.31QYY83 pKa = 11.58LVGAEE88 pKa = 3.64IDD90 pKa = 3.75YY91 pKa = 10.77TEE93 pKa = 4.43GLEE96 pKa = 4.39GSQFVIKK103 pKa = 10.58NPNATTTCGCGSSFSVV119 pKa = 3.54
Molecular weight: 12.81 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D9SHH4|D9SHH4_GALCS Probable chemoreceptor glutamine deamidase CheD OS=Gallionella capsiferriformans (strain ES-2) OX=395494 GN=cheD PE=3 SV=1
MM1 pKa = 7.72RR2 pKa = 11.84LSIIIPILNEE12 pKa = 3.8AVQLPDD18 pKa = 4.63LFAHH22 pKa = 7.1LLPFHH27 pKa = 7.09RR28 pKa = 11.84AGCEE32 pKa = 3.6IIFADD37 pKa = 4.68GGSTDD42 pKa = 3.72GSSKK46 pKa = 10.29LAAVMGFKK54 pKa = 10.22VVQTEE59 pKa = 4.13RR60 pKa = 11.84GRR62 pKa = 11.84ARR64 pKa = 11.84QMNAGAAVAQGNVLLFLHH82 pKa = 7.28ADD84 pKa = 3.09TRR86 pKa = 11.84LPVGAMQQIEE96 pKa = 4.24HH97 pKa = 7.18AFTGPKK103 pKa = 8.86SWGRR107 pKa = 11.84FDD109 pKa = 3.72VCITGRR115 pKa = 11.84PFMLRR120 pKa = 11.84VIGQLMNWRR129 pKa = 11.84SRR131 pKa = 11.84LTGIATGDD139 pKa = 3.1QAIFMRR145 pKa = 11.84RR146 pKa = 11.84IVFEE150 pKa = 4.13RR151 pKa = 11.84LRR153 pKa = 11.84GYY155 pKa = 9.79PDD157 pKa = 3.29QPLMEE162 pKa = 4.94DD163 pKa = 3.25VEE165 pKa = 4.41MCKK168 pKa = 10.16RR169 pKa = 11.84LKK171 pKa = 9.94TFSRR175 pKa = 11.84PACVAHH181 pKa = 6.7CVTTSGRR188 pKa = 11.84RR189 pKa = 11.84WEE191 pKa = 4.09MRR193 pKa = 11.84GVWSTILLMWRR204 pKa = 11.84LRR206 pKa = 11.84WAYY209 pKa = 9.11WRR211 pKa = 11.84GTDD214 pKa = 3.3ATEE217 pKa = 4.09LARR220 pKa = 11.84LYY222 pKa = 10.69RR223 pKa = 4.66
MM1 pKa = 7.72RR2 pKa = 11.84LSIIIPILNEE12 pKa = 3.8AVQLPDD18 pKa = 4.63LFAHH22 pKa = 7.1LLPFHH27 pKa = 7.09RR28 pKa = 11.84AGCEE32 pKa = 3.6IIFADD37 pKa = 4.68GGSTDD42 pKa = 3.72GSSKK46 pKa = 10.29LAAVMGFKK54 pKa = 10.22VVQTEE59 pKa = 4.13RR60 pKa = 11.84GRR62 pKa = 11.84ARR64 pKa = 11.84QMNAGAAVAQGNVLLFLHH82 pKa = 7.28ADD84 pKa = 3.09TRR86 pKa = 11.84LPVGAMQQIEE96 pKa = 4.24HH97 pKa = 7.18AFTGPKK103 pKa = 8.86SWGRR107 pKa = 11.84FDD109 pKa = 3.72VCITGRR115 pKa = 11.84PFMLRR120 pKa = 11.84VIGQLMNWRR129 pKa = 11.84SRR131 pKa = 11.84LTGIATGDD139 pKa = 3.1QAIFMRR145 pKa = 11.84RR146 pKa = 11.84IVFEE150 pKa = 4.13RR151 pKa = 11.84LRR153 pKa = 11.84GYY155 pKa = 9.79PDD157 pKa = 3.29QPLMEE162 pKa = 4.94DD163 pKa = 3.25VEE165 pKa = 4.41MCKK168 pKa = 10.16RR169 pKa = 11.84LKK171 pKa = 9.94TFSRR175 pKa = 11.84PACVAHH181 pKa = 6.7CVTTSGRR188 pKa = 11.84RR189 pKa = 11.84WEE191 pKa = 4.09MRR193 pKa = 11.84GVWSTILLMWRR204 pKa = 11.84LRR206 pKa = 11.84WAYY209 pKa = 9.11WRR211 pKa = 11.84GTDD214 pKa = 3.3ATEE217 pKa = 4.09LARR220 pKa = 11.84LYY222 pKa = 10.69RR223 pKa = 4.66
Molecular weight: 25.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
938798 |
30 |
2854 |
329.1 |
36.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.368 ± 0.064 | 1.07 ± 0.016 |
5.346 ± 0.036 | 5.954 ± 0.048 |
3.772 ± 0.028 | 7.153 ± 0.041 |
2.398 ± 0.023 | 5.83 ± 0.034 |
4.454 ± 0.038 | 10.902 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.728 ± 0.027 | 3.592 ± 0.032 |
4.142 ± 0.032 | 4.267 ± 0.044 |
5.877 ± 0.038 | 6.093 ± 0.035 |
5.07 ± 0.032 | 7.096 ± 0.041 |
1.228 ± 0.02 | 2.658 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
