
Labrenzia alexandrii
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Stappiaceae; Labrenzia
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
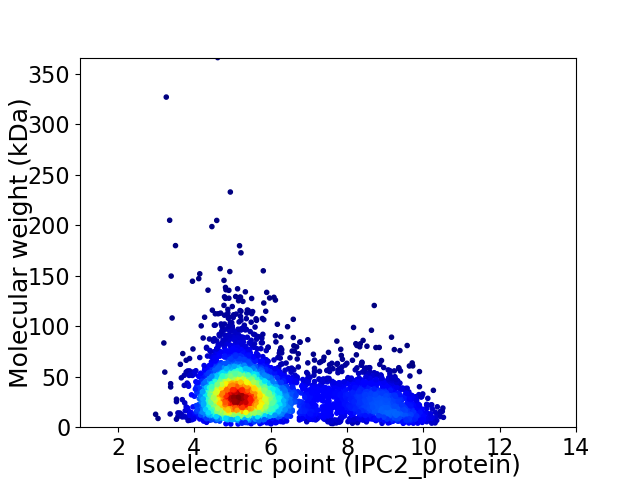
Virtual 2D-PAGE plot for 4993 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0M7AGQ4|A0A0M7AGQ4_9RHOB Uncharacterized protein OS=Labrenzia alexandrii OX=388408 GN=LAX5112_03825 PE=4 SV=1
MM1 pKa = 7.89ADD3 pKa = 3.53NSDD6 pKa = 3.69FHH8 pKa = 5.82QTGDD12 pKa = 4.45EE13 pKa = 4.93DD14 pKa = 6.41DD15 pKa = 3.85PTQNGNGEE23 pKa = 4.17AEE25 pKa = 4.03EE26 pKa = 5.42AIAQLFDD33 pKa = 4.93DD34 pKa = 4.46YY35 pKa = 11.8QSGFNDD41 pKa = 2.99YY42 pKa = 11.51DD43 pKa = 3.32MEE45 pKa = 5.04RR46 pKa = 11.84ICDD49 pKa = 3.94CFALPTIIWQHH60 pKa = 4.99EE61 pKa = 4.24KK62 pKa = 10.88GHH64 pKa = 5.72VFNDD68 pKa = 3.56DD69 pKa = 3.36EE70 pKa = 4.25EE71 pKa = 6.21LIEE74 pKa = 4.69NIEE77 pKa = 4.24VLLKK81 pKa = 10.68ALEE84 pKa = 4.29KK85 pKa = 10.77EE86 pKa = 4.8GVSHH90 pKa = 7.61SDD92 pKa = 3.42FQVVSSHH99 pKa = 5.33VSGPTALVTLDD110 pKa = 3.75WSQEE114 pKa = 4.24SADD117 pKa = 3.74GEE119 pKa = 4.51AVFEE123 pKa = 4.4FTCHH127 pKa = 5.13YY128 pKa = 10.88QLIQDD133 pKa = 4.04GQDD136 pKa = 2.53WMIAGIVNEE145 pKa = 4.14
MM1 pKa = 7.89ADD3 pKa = 3.53NSDD6 pKa = 3.69FHH8 pKa = 5.82QTGDD12 pKa = 4.45EE13 pKa = 4.93DD14 pKa = 6.41DD15 pKa = 3.85PTQNGNGEE23 pKa = 4.17AEE25 pKa = 4.03EE26 pKa = 5.42AIAQLFDD33 pKa = 4.93DD34 pKa = 4.46YY35 pKa = 11.8QSGFNDD41 pKa = 2.99YY42 pKa = 11.51DD43 pKa = 3.32MEE45 pKa = 5.04RR46 pKa = 11.84ICDD49 pKa = 3.94CFALPTIIWQHH60 pKa = 4.99EE61 pKa = 4.24KK62 pKa = 10.88GHH64 pKa = 5.72VFNDD68 pKa = 3.56DD69 pKa = 3.36EE70 pKa = 4.25EE71 pKa = 6.21LIEE74 pKa = 4.69NIEE77 pKa = 4.24VLLKK81 pKa = 10.68ALEE84 pKa = 4.29KK85 pKa = 10.77EE86 pKa = 4.8GVSHH90 pKa = 7.61SDD92 pKa = 3.42FQVVSSHH99 pKa = 5.33VSGPTALVTLDD110 pKa = 3.75WSQEE114 pKa = 4.24SADD117 pKa = 3.74GEE119 pKa = 4.51AVFEE123 pKa = 4.4FTCHH127 pKa = 5.13YY128 pKa = 10.88QLIQDD133 pKa = 4.04GQDD136 pKa = 2.53WMIAGIVNEE145 pKa = 4.14
Molecular weight: 16.3 kDa
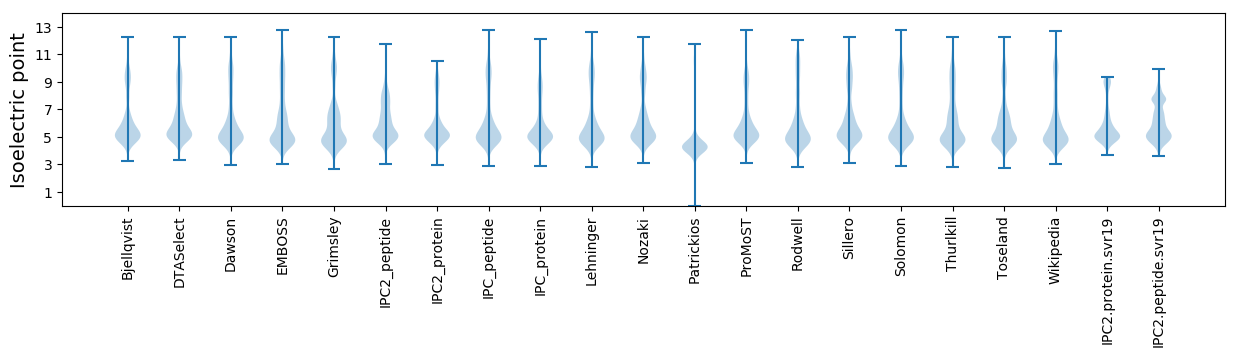
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M7APZ5|A0A0M7APZ5_9RHOB (S)-sulfolactate dehydrogenase OS=Labrenzia alexandrii OX=388408 GN=slcC PE=3 SV=1
MM1 pKa = 7.72RR2 pKa = 11.84FAFIAEE8 pKa = 4.23NQGVLPASRR17 pKa = 11.84LCQIMNVSPRR27 pKa = 11.84GYY29 pKa = 9.65RR30 pKa = 11.84SWRR33 pKa = 11.84DD34 pKa = 3.07RR35 pKa = 11.84PVSDD39 pKa = 3.91RR40 pKa = 11.84QRR42 pKa = 11.84KK43 pKa = 8.95DD44 pKa = 3.45LVVLAHH50 pKa = 6.47IRR52 pKa = 11.84EE53 pKa = 4.12QFRR56 pKa = 11.84LSLGSYY62 pKa = 8.66GRR64 pKa = 11.84PRR66 pKa = 11.84MTEE69 pKa = 3.72EE70 pKa = 4.02LKK72 pKa = 11.09EE73 pKa = 4.02LGLDD77 pKa = 2.92IGHH80 pKa = 6.48RR81 pKa = 11.84RR82 pKa = 11.84VGRR85 pKa = 11.84LMRR88 pKa = 11.84EE89 pKa = 3.46NAITVKK95 pKa = 10.59GNKK98 pKa = 8.97KK99 pKa = 10.19FKK101 pKa = 10.19ATTDD105 pKa = 3.19SNHH108 pKa = 6.68RR109 pKa = 11.84FNIAPNLLKK118 pKa = 10.24RR119 pKa = 11.84DD120 pKa = 4.65FSADD124 pKa = 3.25RR125 pKa = 11.84PNRR128 pKa = 11.84KK129 pKa = 7.97WAGDD133 pKa = 3.22ISYY136 pKa = 10.33VWTRR140 pKa = 11.84EE141 pKa = 3.36GWLYY145 pKa = 10.97LAVILDD151 pKa = 3.56LHH153 pKa = 5.84SRR155 pKa = 11.84RR156 pKa = 11.84VIGWAVSNRR165 pKa = 11.84LKK167 pKa = 10.6RR168 pKa = 11.84DD169 pKa = 3.26LAIRR173 pKa = 11.84ALNMAIALRR182 pKa = 11.84RR183 pKa = 11.84PPKK186 pKa = 10.5GCIHH190 pKa = 5.84HH191 pKa = 6.77TDD193 pKa = 3.63RR194 pKa = 11.84GSQYY198 pKa = 10.54CSHH201 pKa = 8.02DD202 pKa = 3.63YY203 pKa = 10.89QKK205 pKa = 11.49LLRR208 pKa = 11.84QHH210 pKa = 6.85GFQISMSGTGNCFDD224 pKa = 3.78NAAAEE229 pKa = 4.54TFFKK233 pKa = 10.31TIKK236 pKa = 10.85AEE238 pKa = 4.19LLWQKK243 pKa = 10.29SWACRR248 pKa = 11.84RR249 pKa = 11.84EE250 pKa = 3.72AEE252 pKa = 4.19MAIFEE257 pKa = 4.85YY258 pKa = 10.61INGFYY263 pKa = 10.56NPRR266 pKa = 11.84RR267 pKa = 11.84RR268 pKa = 11.84HH269 pKa = 4.97SALAA273 pKa = 3.76
MM1 pKa = 7.72RR2 pKa = 11.84FAFIAEE8 pKa = 4.23NQGVLPASRR17 pKa = 11.84LCQIMNVSPRR27 pKa = 11.84GYY29 pKa = 9.65RR30 pKa = 11.84SWRR33 pKa = 11.84DD34 pKa = 3.07RR35 pKa = 11.84PVSDD39 pKa = 3.91RR40 pKa = 11.84QRR42 pKa = 11.84KK43 pKa = 8.95DD44 pKa = 3.45LVVLAHH50 pKa = 6.47IRR52 pKa = 11.84EE53 pKa = 4.12QFRR56 pKa = 11.84LSLGSYY62 pKa = 8.66GRR64 pKa = 11.84PRR66 pKa = 11.84MTEE69 pKa = 3.72EE70 pKa = 4.02LKK72 pKa = 11.09EE73 pKa = 4.02LGLDD77 pKa = 2.92IGHH80 pKa = 6.48RR81 pKa = 11.84RR82 pKa = 11.84VGRR85 pKa = 11.84LMRR88 pKa = 11.84EE89 pKa = 3.46NAITVKK95 pKa = 10.59GNKK98 pKa = 8.97KK99 pKa = 10.19FKK101 pKa = 10.19ATTDD105 pKa = 3.19SNHH108 pKa = 6.68RR109 pKa = 11.84FNIAPNLLKK118 pKa = 10.24RR119 pKa = 11.84DD120 pKa = 4.65FSADD124 pKa = 3.25RR125 pKa = 11.84PNRR128 pKa = 11.84KK129 pKa = 7.97WAGDD133 pKa = 3.22ISYY136 pKa = 10.33VWTRR140 pKa = 11.84EE141 pKa = 3.36GWLYY145 pKa = 10.97LAVILDD151 pKa = 3.56LHH153 pKa = 5.84SRR155 pKa = 11.84RR156 pKa = 11.84VIGWAVSNRR165 pKa = 11.84LKK167 pKa = 10.6RR168 pKa = 11.84DD169 pKa = 3.26LAIRR173 pKa = 11.84ALNMAIALRR182 pKa = 11.84RR183 pKa = 11.84PPKK186 pKa = 10.5GCIHH190 pKa = 5.84HH191 pKa = 6.77TDD193 pKa = 3.63RR194 pKa = 11.84GSQYY198 pKa = 10.54CSHH201 pKa = 8.02DD202 pKa = 3.63YY203 pKa = 10.89QKK205 pKa = 11.49LLRR208 pKa = 11.84QHH210 pKa = 6.85GFQISMSGTGNCFDD224 pKa = 3.78NAAAEE229 pKa = 4.54TFFKK233 pKa = 10.31TIKK236 pKa = 10.85AEE238 pKa = 4.19LLWQKK243 pKa = 10.29SWACRR248 pKa = 11.84RR249 pKa = 11.84EE250 pKa = 3.72AEE252 pKa = 4.19MAIFEE257 pKa = 4.85YY258 pKa = 10.61INGFYY263 pKa = 10.56NPRR266 pKa = 11.84RR267 pKa = 11.84RR268 pKa = 11.84HH269 pKa = 4.97SALAA273 pKa = 3.76
Molecular weight: 31.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1582945 |
29 |
3368 |
317.0 |
34.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.171 ± 0.043 | 0.867 ± 0.011 |
6.068 ± 0.033 | 6.11 ± 0.037 |
4.03 ± 0.022 | 8.272 ± 0.03 |
1.979 ± 0.018 | 5.517 ± 0.028 |
3.947 ± 0.028 | 10.013 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.549 ± 0.019 | 3.028 ± 0.023 |
4.824 ± 0.028 | 3.358 ± 0.021 |
5.911 ± 0.035 | 5.863 ± 0.023 |
5.62 ± 0.027 | 7.26 ± 0.027 |
1.27 ± 0.012 | 2.341 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
